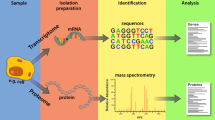
Abstract
Atrial fibrillation (AF) is strongly associated with risk of stroke and heart failure. AF promotes atrial remodeling that increases risk of stroke due to left atrial thrombogenesis, and increases energy demand to support high rate electrical activity and muscle contraction. While many transcriptomic studies have assessed AF-related changes in mRNA abundance, fewer studies have assessed proteomic changes. We performed a proteomic analysis on left atrial appendage (LAA) tissues from 12 patients with a history of AF undergoing elective surgery; atrial rhythm was documented at time of surgery. Proteomic analysis was performed using liquid chromatography with mass spectrometry (LC/MS-MS). Data-dependent analysis identified 3090 unique proteins, with 408 differentially expressed between sinus rhythm and AF. Ingenuity Pathway Analysis of differentially expressed proteins identified mitochondrial dysfunction, oxidative phosphorylation, and sirtuin signaling among the most affected pathways. Increased abundance of electron transport chain (ETC) proteins in AF was accompanied by decreased expression of ETC complex assembly factors, tricarboxylic acid cycle proteins, and other key metabolic modulators. Discordant changes were also evident in the contractile unit with both up and downregulation of key components. Similar pathways were affected in a comparison of patients with a history of persistent vs. paroxysmal AF, presenting for surgery in sinus rhythm. Together, these data suggest that while the LAA attempts to meet the energetic demands of AF, an uncoordinated response may reduce ATP availability, contribute to tissue contractile and electrophysiologic heterogeneity, and promote a progression of AF from paroxysmal episodes to development of a substrate amenable to persistent arrhythmia.






Similar content being viewed by others
Data availability
Full summary data are included in the manuscript and the accompanying online resources. Proteomic data have been uploaded to the PRIDE database.
References
Barth AS, Kumordzie A, Tomaselli GF (2016) Orchestrated regulation of energy supply and energy expenditure: transcriptional coexpression of metabolism, ion homeostasis, and sarcomeric genes in mammalian myocardium. Heart Rhythm 13:1131–1139. https://doi.org/10.1016/j.hrthm.2016.01.009
Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kaab S, Hinterseer M, Kartmann H, Kreuzer E, Dugas M, Steinbeck G, Nabauer M (2005) Reprogramming of the human atrial transcriptome in permanent atrial fibrillation: expression of a ventricular-like genomic signature. Circ Res 96:1022–1029. https://doi.org/10.1161/01.RES.0000165480.82737.33
Barth AS, Merk S, Arnoldi E, Zwermann L, Kloos P, Gebauer M, Steinmeyer K, Bleich M, Kaab S, Pfeufer A, Uberfuhr P, Dugas M, Steinbeck G, Nabauer M (2005) Functional profiling of human atrial and ventricular gene expression. Pflugers Arch 450:201–208. https://doi.org/10.1007/s00424-005-1404-8
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol 57:289–300
Calvo SE, Clauser KR, Mootha VK (2016) MitoCarta2.0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44:D1251–D1257. https://doi.org/10.1093/nar/gkv1003
Cardin S, Libby E, Pelletier P, Le Bouter S, Shiroshita-Takeshita A, Le Meur N, Leger J, Demolombe S, Ponton A, Glass L, Nattel S (2007) Contrasting gene expression profiles in two canine models of atrial fibrillation. Circ Res 100:425–433. https://doi.org/10.1161/01.RES.0000258428.09589.1a
Carnes CA, Janssen PM, Ruehr ML, Nakayama H, Nakayama T, Haase H, Bauer JA, Chung MK, Fearon IM, Gillinov AM, Hamlin RL, Van Wagoner DR (2007) Atrial glutathione content, calcium current, and contractility. J Biol Chem 282:28063–28073. https://doi.org/10.1074/jbc.M704893200
Chan MY, Lin M, Lucas J, Moseley A, Thompson JW, Cyr D, Ueda H, Kajikawa M, Ortel TL, Becker RC (2012) Plasma proteomics of patients with non-valvular atrial fibrillation on chronic anti-coagulation with warfarin or a direct factor Xa inhibitor. Thromb Haemost 108:1180–1191. https://doi.org/10.1160/TH12-05-0310
Chen J, Behnam E, Huang J, Moffatt MF, Schaid DJ, Liang L, Lin X (2017) Fast and robust adjustment of cell mixtures in epigenome-wide association studies with SmartSVA. BMC Genomics 18:413. https://doi.org/10.1186/s12864-017-3808-1
Cox J, Hein MY, Luber CA, Paron I, Nagaraj N, Mann M (2014) Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13:2513–2526. https://doi.org/10.1074/mcp.M113.031591
De Souza AI, Cardin S, Wait R, Chung YL, Vijayakumar M, Maguy A, Camm AJ, Nattel S (2010) Proteomic and metabolomic analysis of atrial profibrillatory remodelling in congestive heart failure. J Mol Cell Cardiol 49:851–863. https://doi.org/10.1016/j.yjmcc.2010.07.008
Deshmukh A, Barnard J, Sun H, Newton D, Castel L, Pettersson G, Johnston D, Roselli E, Gillinov AM, McCurry K, Moravec C, Smith JD, Van Wagoner DR, Chung MK (2015) Left atrial transcriptional changes associated with atrial fibrillation susceptibility and persistence. Circ Arrhythm Electrophysiol 8:32–41. https://doi.org/10.1161/CIRCEP.114.001632
Dinanian S, Boixel C, Juin C, Hulot JS, Coulombe A, Rucker-Martin C, Bonnet N, Le Grand B, Slama M, Mercadier JJ, Hatem SN (2008) Downregulation of the calcium current in human right atrial myocytes from patients in sinus rhythm but with a high risk of atrial fibrillation. Eur Heart J 29:1190–1197. https://doi.org/10.1093/eurheartj/ehn140
Doll S, Dressen M, Geyer PE, Itzhak DN, Braun C, Doppler SA, Meier F, Deutsch MA, Lahm H, Lange R, Krane M, Mann M (2017) Region and cell-type resolved quantitative proteomic map of the human heart. Nat Commun 8:1469. https://doi.org/10.1038/s41467-017-01747-2
Goudarzi M, Ross MM, Zhou W, Van Meter A, Deng J, Martin LM, Martin C, Liotta L, Petricoin E, Ad N (2011) Development of a novel proteomic approach for mitochondrial proteomics from cardiac tissue from patients with atrial fibrillation. J Proteome Res 10:3484–3492. https://doi.org/10.1021/pr200108m
Harada M, Tadevosyan A, Qi X, Xiao J, Liu T, Voigt N, Karck M, Kamler M, Kodama I, Murohara T, Dobrev D, Nattel S (2015) Atrial fibrillation activates AMP-dependent protein kinase and its regulation of cellular calcium handling: potential role in metabolic adaptation and prevention of progression. J Am Coll Cardiol 66:47–58. https://doi.org/10.1016/j.jacc.2015.04.056
Heijman J, Abdoust PE, Voigt N, Nattel S, Dobrev D (2015) Computational models of atrial cellular electrophysiology and calcium handling, and their role in atrial fibrillation. J Physiol 594:537–553. https://doi.org/10.1113/JP271404
Hool LC (2009) The L-type Ca(2+) channel as a potential mediator of pathology during alterations in cellular redox state. Heart Lung Circ 18:3–10. https://doi.org/10.1016/j.hlc.2008.11.004
Hsu J, Gore-Panter S, Tchou G, Castel L, Lovano B, Moravec CS, Pettersson GB, Roselli EE, Gillinov AM, McCurry KR, Smedira NG, Barnard J, Van Wagoner DR, Chung MK, Smith JD (2018) Genetic control of left atrial gene expression yields insights into the genetic susceptibility for atrial fibrillation. Circ Genom Precis Med 11:e002107. https://doi.org/10.1161/CIRCGEN.118.002107
Klein O, Hanke T, Nebrich G, Yan J, Schubert B, Giavalisco P, Noack F, Thiele H, Mohamed SA (2018) Imaging mass spectrometry for characterization of atrial fibrillation subtypes. Proteomics Clin Appl 12:e1700155. https://doi.org/10.1002/prca.201700155
Ko D, Benson MD, Ngo D, Yang Q, Larson MG, Wang TJ, Trinquart L, McManus DD, Lubitz SA, Ellinor PT, Vasan RS, Gerszten RE, Benjamin EJ, Lin H (2019) Proteomics profiling and risk of new-onset atrial fibrillation: Framingham Heart Study. J Am Heart Assoc 8:e010976. https://doi.org/10.1161/JAHA.118.010976
Kornej J, Buttner P, Hammer E, Engelmann B, Dinov B, Sommer P, Husser D, Hindricks G, Volker U, Bollmann A (2018) Circulating proteomic patterns in AF related left atrial remodeling indicate involvement of coagulation and complement cascade. PLoS One 13:e0198461. https://doi.org/10.1371/journal.pone.0198461
Kourliouros A, Yin X, Didangelos A, Hosseini MT, Valencia O, Mayr M, Jahangiri M (2011) Substrate modifications precede the development of atrial fibrillation after cardiac surgery: a proteomic study. Ann Thorac Surg 92:104–110. https://doi.org/10.1016/j.athoracsur.2011.03.071
Kraja AT, Liu C, Fetterman JL, Graff M, Have CT, Gu C, Yanek LR, Feitosa MF, Arking DE, Chasman DI, Young K, Ligthart S, Hill WD, Weiss S, Luan J, Giulianini F, Li-Gao R, Hartwig FP, Lin SJ, Wang L, Richardson TG, Yao J, Fernandez EP, Ghanbari M, Wojczynski MK, Lee WJ, Argos M, Armasu SM, Barve RA, Ryan KA, An P, Baranski TJ, Bielinski SJ, Bowden DW, Broeckel U, Christensen K, Chu AY, Corley J, Cox SR, Uitterlinden AG, Rivadeneira F, Cropp CD, Daw EW, van Heemst D, de Las FL, Gao H, Tzoulaki I, Ahluwalia TS, de Mutsert R, Emery LS, Erzurumluoglu AM, Perry JA, Fu M, Forouhi NG, Gu Z, Hai Y, Harris SE, Hemani G, Hunt SC, Irvin MR, Jonsson AE, Justice AE, Kerrison ND, Larson NB, Lin KH, Love-Gregory LD, Mathias RA, Lee JH, Nauck M, Noordam R, Ong KK, Pankow J, Patki A, Pattie A, Petersmann A, Qi Q, Ribel-Madsen R, Rohde R, Sandow K, Schnurr TM, Sofer T, Starr JM, Taylor AM, Teumer A, Timpson NJ, de Haan HG, Wang Y, Weeke PE, Williams C, Wu H, Yang W, Zeng D, Witte DR, Weir BS, Wareham NJ, Vestergaard H, Turner ST, Torp-Pedersen C, Stergiakouli E, Sheu WH, Rosendaal FR, Ikram MA, Franco OH, Ridker PM, Perls TT, Pedersen O, Nohr EA, Newman AB, Linneberg A, Langenberg C, Kilpelainen TO, Kardia SLR, Jorgensen ME, Jorgensen T, Sorensen TIA, Homuth G, Hansen T, Goodarzi MO, Deary IJ, Christensen C, Chen YI, Chakravarti A, Brandslund I, Bonnelykke K, Taylor KD, Wilson JG, Rodriguez S, Davies G, Horta BL, Thyagarajan B, Rao DC, Grarup N, Davila-Roman VG, Hudson G, Guo X, Arnett DK, Hayward C, Vaidya D, Mook-Kanamori DO, Tiwari HK, Levy D, Loos RJF, Dehghan A, Elliott P, Malik AN, Scott RA, Becker DM, de Andrade M, Province MA, Meigs JB, Rotter JI, North KE (2019) Associations of mitochondrial and nuclear mitochondrial variants and genes with seven metabolic traits. Am J Hum Genet 104:112–138. https://doi.org/10.1016/j.ajhg.2018.12.001
Lamirault G, Gaborit N, Le Meur N, Chevalier C, Lande G, Demolombe S, Escande D, Nattel S, Leger JJ, Steenman M (2006) Gene expression profile associated with chronic atrial fibrillation and underlying valvular heart disease in man. J Mol Cell Cardiol 40:173–184. https://doi.org/10.1016/j.yjmcc.2005.09.004
Lazar C, Gatto L, Ferro M, Bruley C, Burger T (2016) Accounting for the multiple natures of missing values in label-free quantitative proteomics data sets to compare imputation strategies. J Proteome Res 15:1116–1125. https://doi.org/10.1021/acs.jproteome.5b00981
Le Grand BL, Hatem S, Deroubaix E, Couetil JP, Coraboeuf E (1994) Depressed transient outward and calcium currents in dilated human atria. Cardiovasc Res 28:548–556. https://doi.org/10.1093/cvr/28.4.548
Lind L, Sundstrom J, Stenemo M, Hagstrom E, Arnlov J (2017) Discovery of new biomarkers for atrial fibrillation using a custom-made proteomics chip. Heart 103:377–382. https://doi.org/10.1136/heartjnl-2016-309764
Liu JC, Syder NC, Ghorashi NS, Willingham TB, Parks RJ, Sun J, Fergusson MM, Liu J, Holmstrom KM, Menazza S, Springer DA, Liu C, Glancy B, Finkel T, Murphy E (2020) EMRE is essential for mitochondrial calcium uniporter activity in a mouse model. JCI Insight 5. https://doi.org/10.1172/jci.insight.134063
Liu LJ, Yao FJ, Lu GH, Xu CG, Xu Z, Tang K, Cheng YJ, Gao XR, Wu SH (2016) The role of the Rho/ROCK pathway in Ang II and TGF-beta1-induced atrial remodeling. PLoS One 11:e0161625. https://doi.org/10.1371/journal.pone.0161625
Mayr M, Yusuf S, Weir G, Chung YL, Mayr U, Yin X, Ladroue C, Madhu B, Roberts N, De Souza A, Fredericks S, Stubbs M, Griffiths JR, Jahangiri M, Xu Q, Camm AJ (2008) Combined metabolomic and proteomic analysis of human atrial fibrillation. J Am Coll Cardiol 51:585–594. https://doi.org/10.1016/j.jacc.2007.09.055
Meijering RAM, Wiersma M, Zhang D, Lanters EAH, Hoogstra-Berends F, Scholma J, Diks S, Qi X, de Groot NMS, Nattel S, Henning RH, Brundel B (2018) Application of kinomic array analysis to screen for altered kinases in atrial fibrillation remodeling. Heart Rhythm 15:1708–1716. https://doi.org/10.1016/j.hrthm.2018.06.014
Mihm MJ, Yu F, Carnes CA, Reiser PJ, McCarthy PM, Van Wagoner DR, Bauer JA (2001) Impaired myofibrillar energetics and oxidative injury during human atrial fibrillation. Circulation 104:174–180. https://doi.org/10.1161/01.cir.104.2.174
Nielsen JB, Thorolfsdottir RB, Fritsche LG, Zhou W, Skov MW, Graham SE, Herron TJ, McCarthy S, Schmidt EM, Sveinbjornsson G, Surakka I, Mathis MR, Yamazaki M, Crawford RD, Gabrielsen ME, Skogholt AH, Holmen OL, Lin M, Wolford BN, Dey R, Dalen H, Sulem P, Chung JH, Backman JD, Arnar DO, Thorsteinsdottir U, Baras A, O’Dushlaine C, Holst AG, Wen X, Hornsby W, Dewey FE, Boehnke M, Kheterpal S, Mukherjee B, Lee S, Kang HM, Holm H, Kitzman J, Shavit JA, Jalife J, Brummett CM, Teslovich TM, Carey DJ, Gudbjartsson DF, Stefansson K, Abecasis GR, Hveem K, Willer CJ (2018) Biobank-driven genomic discovery yields new insight into atrial fibrillation biology. Nat Genet 50:1234–1239. https://doi.org/10.1038/s41588-018-0171-3
Oropeza-Almazan Y, Blatter LA (2020) Mitochondrial calcium uniporter complex activation protects against calcium alternans in atrial myocytes. American journal of physiology Heart and circulatory physiology 0:null. https://doi.org/10.1152/ajpheart.00375.2020
Rahman F, Kwan GF, Benjamin EJ (2014) Global epidemiology of atrial fibrillation. Nat Rev Cardiol 11:639–654. https://doi.org/10.1038/nrcardio.2014.118
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Roselli C, Chaffin MD, Weng LC, Aeschbacher S, Ahlberg G, Albert CM, Almgren P, Alonso A, Anderson CD, Aragam KG, Arking DE, Barnard J, Bartz TM, Benjamin EJ, Bihlmeyer NA, Bis JC, Bloom HL, Boerwinkle E, Bottinger EB, Brody JA, Calkins H, Campbell A, Cappola TP, Carlquist J, Chasman DI, Chen LY, Chen YI, Choi EK, Choi SH, Christophersen IE, Chung MK, Cole JW, Conen D, Cook J, Crijns HJ, Cutler MJ, Damrauer SM, Daniels BR, Darbar D, Delgado G, Denny JC, Dichgans M, Dorr M, Dudink EA, Dudley SC, Esa N, Esko T, Eskola M, Fatkin D, Felix SB, Ford I, Franco OH, Geelhoed B, Grewal RP, Gudnason V, Guo X, Gupta N, Gustafsson S, Gutmann R, Hamsten A, Harris TB, Hayward C, Heckbert SR, Hernesniemi J, Hocking LJ, Hofman A, Horimoto A, Huang J, Huang PL, Huffman J, Ingelsson E, Ipek EG, Ito K, Jimenez-Conde J, Johnson R, Jukema JW, Kaab S, Kahonen M, Kamatani Y, Kane JP, Kastrati A, Kathiresan S, Katschnig-Winter P, Kavousi M, Kessler T, Kietselaer BL, Kirchhof P, Kleber ME, Knight S, Krieger JE, Kubo M, Launer LJ, Laurikka J, Lehtimaki T, Leineweber K, Lemaitre RN, Li M, Lim HE, Lin HJ, Lin H, Lind L, Lindgren CM, Lokki ML, London B, Loos RJF, Low SK, Lu Y, Lyytikainen LP, Macfarlane PW, Magnusson PK, Mahajan A, Malik R, Mansur AJ, Marcus GM, Margolin L, Margulies KB, Marz W, McManus DD, Melander O, Mohanty S, Montgomery JA, Morley MP, Morris AP, Muller-Nurasyid M, Natale A, Nazarian S, Neumann B, Newton-Cheh C, Niemeijer MN, Nikus K, Nilsson P, Noordam R, Oellers H, Olesen MS, Orho-Melander M, Padmanabhan S, Pak HN, Pare G, Pedersen NL, Pera J, Pereira A, Porteous D, Psaty BM, Pulit SL, Pullinger CR, Rader DJ, Refsgaard L, Ribases M, Ridker PM, Rienstra M, Risch L, Roden DM, Rosand J, Rosenberg MA, Rost N, Rotter JI, Saba S, Sandhu RK, Schnabel RB, Schramm K, Schunkert H, Schurman C, Scott SA, Seppala I, Shaffer C, Shah S, Shalaby AA, Shim J, Shoemaker MB, Siland JE, Sinisalo J, Sinner MF, Slowik A, Smith AV, Smith BH, Smith JG, Smith JD, Smith NL, Soliman EZ, Sotoodehnia N, Stricker BH, Sun A, Sun H, Svendsen JH, Tanaka T, Tanriverdi K, Taylor KD, Teder-Laving M, Teumer A, Theriault S, Trompet S, Tucker NR, Tveit A, Uitterlinden AG, Van Der Harst P, Van Gelder IC, Van Wagoner DR, Verweij N, Vlachopoulou E, Volker U, Wang B, Weeke PE, Weijs B, Weiss R, Weiss S, Wells QS, Wiggins KL, Wong JA, Woo D, Worrall BB, Yang PS, Yao J, Yoneda ZT, Zeller T, Zeng L, Lubitz SA, Lunetta KL, Ellinor PT (2018) Multi-ethnic genome-wide association study for atrial fibrillation. Nat Genet 50:1225–1233. https://doi.org/10.1038/s41588-018-0133-9
Seidl MD, Stein J, Hamer S, Pluteanu F, Scholz B, Wardelmann E, Huge A, Witten A, Stoll M, Hammer E, Volker U, Muller FU (2017) Characterization of the genetic program linked to the development of atrial fibrillation in CREM-IbDeltaC-X Mice. Circ Arrhythm Electrophysiol 10:10. https://doi.org/10.1161/CIRCEP.117.005075
Silva TS, Richard N (2016) Visualization and differential analysis of protein expression data using R. Methods Mol Biol 1362:105–118. https://doi.org/10.1007/978-1-4939-3106-4_6
Skasa M, Jungling E, Picht E, Schondube F, Luckhoff A (2001) L-type calcium currents in atrial myocytes from patients with persistent and non-persistent atrial fibrillation. Basic Res Cardiol 96:151–159. https://doi.org/10.1007/s003950170065
Staerk L, Preis SR, Lin H, Lubitz SA, Ellinor PT, Levy D, Benjamin EJ, Trinquart L (2020) Protein biomarkers and risk of atrial fibrillation: the FHS. Circ Arrhythm Electrophysiol 13:e007607. https://doi.org/10.1161/CIRCEP.119.007607
Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA (2012) Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol 32:1363–1373. https://doi.org/10.1128/MCB.06369-11
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F (2015) Proteomics. Tissue-based map of the human proteome. Science 347:1260419. https://doi.org/10.1126/science.1260419
Vahsen N, Cande C, Briere JJ, Benit P, Joza N, Larochette N, Mastroberardino PG, Pequignot MO, Casares N, Lazar V, Feraud O, Debili N, Wissing S, Engelhardt S, Madeo F, Piacentini M, Penninger JM, Schagger H, Rustin P, Kroemer G (2004) AIF deficiency compromises oxidative phosphorylation. EMBO J 23:4679–4689. https://doi.org/10.1038/sj.emboj.7600461
Valikangas T, Suomi T, Elo LL (2018) A systematic evaluation of normalization methods in quantitative label-free proteomics. Brief Bioinform 19:1–11. https://doi.org/10.1093/bib/bbw095
Van Wagoner DR (2008) Oxidative stress and inflammation in atrial fibrillation: role in pathogenesis and potential as a therapeutic target. J Cardiovasc Pharmacol 52:306–313. https://doi.org/10.1097/FJC.0b013e31817f9398
Van Wagoner DR, Pond AL, Lamorgese M, Rossie SS, McCarthy PM, Nerbonne JM (1999) Atrial L-type Ca2+ currents and human atrial fibrillation. Circ Res 85:428–436. https://doi.org/10.1161/01.res.85.5.428
Wang J, Wang Y, Han J, Li Y, Xie C, Xie L, Shi J, Zhang J, Yang B, Chen D, Meng X (2015) Integrated analysis of microRNA and mRNA expression profiles in the left atrium of patients with nonvalvular paroxysmal atrial fibrillation: Role of miR-146b-5p in atrial fibrosis. Heart Rhythm 12:1018–1026. https://doi.org/10.1016/j.hrthm.2015.01.026
Wiersma M, van Marion DMS, Wust RCI, Houtkooper RH, Zhang D, Groot NMS, Henning RH, Brundel B (2019) Mitochondrial dysfunction underlies cardiomyocyte remodeling in experimental and clinical atrial fibrillation. Cells 8:1202. https://doi.org/10.3390/cells8101202
Yan J, Zhao W, Thomson JK, Gao X, DeMarco DM, Carrillo E, Chen B, Wu X, Ginsburg KS, Bakhos M, Bers DM, Anderson ME, Song LS, Fill M, Ai X (2018) Stress signaling JNK2 crosstalk with CaMKII underlies enhanced atrial arrhythmogenesis. Circ Res 122:821–835. https://doi.org/10.1161/CIRCRESAHA.117.312536
Zhou J, Gao J, Liu Y, Gu S, Zhang X, An X, Yan J, Xin Y, Su P (2014) Human atrium transcript analysis of permanent atrial fibrillation. Int Heart J 55:71–77. https://doi.org/10.1536/ihj.13-196
Acknowledgments
The Fusion Lumos LC MS/MS instrument was purchased via an NIH shared instrument grant, 1S10OD023436-01.
Funding
Funding was provided by a Strategically Focused Research Network in Atrial Fibrillation grant from the American Heart Association (18SFRN34110067, 18SFRN34170442) and by the National Institutes of Health (R01 HL111314).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
All surgical patients provided informed consent for research use of discarded atrial tissues. Prior to 2008, verbal consent was obtained and documented in the patient medical records in a process approved by the Cleveland Clinic Institutional Review Board (IRB, project 18-1501).
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the special issue on Calcium Signal Dynamics in Cardiac Myocytes and Fibroblasts: Mechanisms in Pflügers Archiv—European Journal of Physiology
Supplementary Information
ESM 1
Online Resource 1 3090 proteins with UniProtKB identifiers were suitable for analysis. Previously identified potential AF risk genes detected in our analysis are identified in column J with yes (Y). Proteins are presented in alphabetical order. Online Resource 2 408 proteins were differentially expressed in atrial fibrillation vs. sinus rhythm. Proteins are identified by gene name and UniProtKB identifier. Online Resource 3 362 Canonical Pathways identified by Ingenuity Pathway Analysis. Pathway name, false discovery rate (-log(p-value)), and proteins that were differentially expressed in each pathway are identified. Pathways are presented in order of false discovery rate, with the most significant pathway listed first. Online Resource 4 Top upstream regulators identified by Ingenuity Pathway Analysis (IPA) of proteins that were differentially expressed in our analysis. Upstream regulators are predicted by IPA using previously published literature. Some of these upstream regulators were themselves proteins that were differentially expressed in our dataset and therefore the expression log ratio, a measure of fold change (column B), is included. Differentially expressed proteins that are predicted to be influenced by each upstream regulator are designated. Significance was determined using a false discovery rate p < 0.05. Upstream regulators are presented in order of false discovery rate, with the most significant upstream regulator listed first. Online Resource 5 Differentially expressed proteins in left atrial appendage of patients in sinus rhythm at the time of tissue acquisition, but with a clinical history of either paroxysmal (n = 4) or persistent AF (n = 4) (p < 0.05). Proteins are identified by gene name and UniProtKB identifier. Online Resource 6 Canonical Pathways identified by Ingenuity Pathway Analysis in patients in sinus rhythm at the time of tissue acquisition, but with a clinical history of either paroxysmal or persistent AF. Pathway name, false discovery rate (-log(p-value)), and proteins that were differentially expressed in each pathway are identified. Pathways are presented in order of false discovery rate, with the most significant pathway listed first. (XLSX 816 kb). Online Resource 7 Top upstream regulators identified by Ingenuity Pathway Analysis (IPA) of proteins that were differentially expressed in patients in sinus rhythm at the time of tissue acquisition, but with a clinical history of either paroxysmal or persistent AF. Upstream regulators are predicted by IPA using previously published literature. Some of these upstream regulators were themselves proteins that were differentially expressed in our dataset and therefore the expression log ratio, a measure of fold change (column B), is included. Differentially expressed proteins that are predicted to be influenced by each upstream regulator are designated. Significance was determined using a false discovery rate p < 0.05. Upstream regulators are presented in order of false discovery rate, with the most significant upstream regulator listed first
ESM 2
Supplemental Fig. 1 Proteins differentially expressed in patients in sinus rhythm at the time of tissue acquisition, but with a clinical history of either paroxysmal or persistent AF. Upregulated proteins are designated red, downregulated are designated green. Abbreviations used: adenylate kinase (AK4), armadillo repeat containing X-linked 1 (ARMCX1), aspartate β-hydroxylase (ASPH), calcyclin binding protein (CACYBP), enoyl-CoA δ isomerase 1 (ECI1), filamin C (FLNC), heterogeneous nuclear ribonucleoprotein A/B (HNRNPAB), mitochondrial ribosomal protein 47 (MRPL47), mitochondrial translation initiation factor (MTIF), NADH:ubiquinone oxidoreductase subunit A7 (NDUFA7), nipsnap homolog 3B (NIPSNAP3B), nuclear transport factor 2 (NUTF2), peptidylprolyl isomerase D (PPID), S100 calcium binding protein A8 (S100A8), splicing factor 3B subunit 1 (SF3B1), tubulin polymerization promoting protein family member 3 (TPPP3), triadin (TRDN), thyroid hormone receptor interactor 11 (TRIP11), and tryptophanyl TRNA synthetase 2, mitochondrial (WARS2). Supplemental Fig. 2 Top 35 canonical pathways identified by Ingenuity Pathway Analysis of proteins differentially expressed in patients in sinus rhythm at the time of tissue acquisition, but with a clinical history of either paroxysmal or persistent AF. Pathways are presented in order of significance, with the most significant pathway listed first. Significance was determined using a false discovery rate p < 0.05. Pathways that are predicted to be activated are designated in orange shades. Pathways that predicted to be inhibited are designated in blue shades. Gray designates pathways for which there was not sufficient information to predict directionality. (PDF 277 kb)
Rights and permissions
About this article
Cite this article
Rennison, J.H., Li, L., Lin, C.R. et al. Atrial fibrillation rhythm is associated with marked changes in metabolic and myofibrillar protein expression in left atrial appendage. Pflugers Arch - Eur J Physiol 473, 461–475 (2021). https://doi.org/10.1007/s00424-021-02514-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-021-02514-5