Abstract
The inhibition of the human ether-à-go-go-related (hERG) K+ channels is the major cause of long QT syndromes inducing fatal cardiac arrhythmias. Ergtoxin 1 (ErgTx1) belongs to scorpion-toxins, which are K+ channel-blockers, and binds to hERG channel with 1:1 stoichiometry and high affinity (K d ∼ 10 nM). Nevertheless, patch-clamp recordings recently demonstrated that ErgTx1 does not establish complete blockade of hERG currents, even at high ErgTx1 concentrations. Such phenomenon is supposed to be consistent with highly dynamic conformational changes of the outer pore domain of hERG. In this study, simultaneous topography and recognition imaging (TREC) on hERG HEK 293 cells was used to visualize binding sites on the extracellular part of hERG channel (on S1–S2 region) for Anti-Kv11.1 (hERG-extracellular-antibody). The recognition maps of hERG channels contained recognition spots, haphazardly distributed and organized in clusters. Recognition images after the addition of ErgTx1 at high concentrations (∼1 μM) revealed subsequent partial disappearance of clusters, indicating that ErgTx1 was bound to the S1–S2 region. These results were supported by AFM force spectroscopy data, showing for the first time that voltage sensing domain (S1–S4) of hERG K+ channel might be one of the multiple binding sites of ErgTx1.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human ether-à-go-go-related gene (hERG) potassium (K+) channel plays important roles in heart [1], peripheral sympathetic ganglia, brain, and tumor cells. hERG K+ channels mediate the rapidly activating delayed rectifier K+ current (I Kr) [2] that plays a key role in repolarization of the ventricular action potential [3, 4]. hERG channels [5, 6] are largely involved in myocardial repolarization [2, 7, 8] and associated with both the inherited and the acquired (drug-induced) long QT syndromes (LQT2) that may be responsible of fatal cardiac arrhythmias. The inherited mutations in hERG are adverse drug effects that suppress I Kr function. hERG expresses in noncardiac cell types [9] and plays significant role under physiological or pathological conditions [10]. All these findings indicate that agents that could suppress or enhance the hERG channel function would find important therapeutic applications in other fields then antiarrhythmic therapy [11]. Therefore, it is of great consequence to get structural information about the hERG domains (voltage sensor and pore domains (Fig. 1)), which are involved in determining drug binding affinity and specificity. In analogy to the Kv family of K+ channels, hERG comprises a tetramer, with each subunit containing six transmembrane helices (Fig. 1) [5]. hERG K+ channels are unusually promiscuous drug target through internal pore blockade [2, 12] and are especially sensitive to external cations acting as a gating modifiers [13–17].
Schematic representation of the hERG subunit. One subunit contains six α-helical transmembrane helices S1–S6 and comprises two functionally distinct modules, one that senses transmembrane potential, voltage-sensor domain (S1–S4) and one that forms the K+-selective pore (S5–S6). S4 domain contains multiple basic (+) amino acids and acidic Asp residues (−) in S1–S3 that can form salt bridges with specific basic residues in S4 during gating. Movements of the voltage-sensor domain enable the pore domain to open and close in response to changes in membrane potential. The pore domain contains the highly conserved K+ channel features—the pore helix and selectivity filter, which permit selective passage of K+ ions
Peptide toxins can bind to the outer vestibule of the hERG channel [18, 19] and thus are very useful tool for understanding the structure of hERG unit (Fig. 1). They are found in a large variety of venoms such as scorpions, snakes, bees, sea anemone, and spiders and have been widely used for structural analyses, localization, and determination of pore forming regions of voltage-dependent K+ channels [1]. Toxins modulating K+ channel function are short-chain peptides with a length of ∼30–40 aa and contain three or four disulfide bonds [20]. Importantly, many of the structural constraints of pore-blocking toxins have been gained from the crystal structure of the KcsA K+ channel [21]. Unlike the pore-blocking toxins, which typically bind to the outer pore domain, the location and the structural determinants of receptors for gating modifier toxins are poorly elucidated. Gating-modifier toxins typically bind to the voltage-sensor domain (i.e. S1–S4 domains) and shift the voltage range for channel activation [22]. Thus, the gating modifier toxins represent ideal probes to examine the structure of gating domains (S1–S4). Some hERG-specific peptide toxins have been recently identified: peptide (42 aa) isolated from the Mexican scorpion Centruroides noxius (named ergtoxin 1 (ErgTx1)) [1], toxin (36 aa) from the scorpion Buthus eupeus (BeKm-1) [23], ErgTx2 [24], peptide (43 aa) from the American scorpion Centruroides sculpturatus (CsEKerg1) [25], and saxitoxin (STX) [22].
In this study, we focused on the interaction of ErgTx1, which is entirely different from 10 other subfamilies of known scorpion toxins [1], with the wild-type hERG K+ channel. Peptide toxins usually block the pore of the channel, either directly by occupying the selective filter or by binding to an electrostatic ring surrounding the pore. Previously, it has been identified that ErgTx1 binds to the outer vestibule of the hERG channel [26]. The binding site is probably formed by uncharged residues in S5-P and P-S6 linkers of the hERG channel [27]). A characteristic feature of the action of ErgTx1 on hERG is incomplete block of macroscopic current event at concentrations orders-of-magnitude higher than the K d value. Such effect suggests that ErgTx1 is a gating modifier rather than a pore blocker [27, 28]. In addition, it binds near the pore and can not fully occlude the permeation pathway [29, 30]. The binding site for ErgTx1 on hERG is thought to be formed, at least in part, by the extracellular linker between S5 transmembrane helix and the pore helix (S5P linker) [27], which is critically involved in voltage-dependent inactivation in hERG [31].
Generally, the information about the structure and function of different voltage-gated channels in living cells (including hERG K+ channels) were gained from patch-clamp investigations. Here, we introduce an alternative way to probe the surface topology of hERG channel by using AFM (dynamic recognition imaging and single molecule force spectroscopy). Due to continuous progress in the technical aspects of the AFM and tip functionalization procedures, the investigations of receptor–ligand interactions on living cells at the single-molecule level have become possible [32–34]. To localize receptor–ligand recognition in vitro systems and in living cells, fluorescence techniques (immunochemistry [35] or single molecule optical microscopy [36] are commonly used. However, due to the limited resolution (from few tens to 200 nm), the recognition sites cannot be resolved on the nanometer scale nor can they be correlated to topography features. Combination of high-resolution atomic force microscope (AFM) topography imaging with single molecule force spectroscopy provides a unique possibility for the detection of specific molecular recognition events. With the recent development of simultaneous topography and recognition imaging (TREC) [37–39] it becomes possible to quickly obtain the local distribution of receptors on cell surface with unprecedented lateral resolution of 5 nm [40]. At present, no other microscopic techniques are able to provide directly both structural information of a biological sample and related functional information at such high spatial resolution.
In this study, we have used dynamic recognition imaging (TREC) with AFM force spectroscopy to locally identify a new receptor site(s) for ErgTx1 in voltage sensing domain of hERG K+ channel with a goal to extend our understanding of microscopic mechanism by which ErgTx1 blocks K+ channels.
Materials and methods
Cell culture
Experiments were performed either on wild-type human embryonic kidney (HEK-293) cells or on HEK-293 cells stably expressing wild-type hERG (abbreviated as hERG HEK-293) (kindly provided by Professor Craig January (University of Wisconsin, Madison, WI, USA). HEK-293 cells were grown in Dulbecco’s modified Eagles medium (DMEM), supplemented with 50 U/ml penicillin-G, 50 μg streptomycin, and 10% fetal calf serum (FCS) in a humidified atmosphere (5% CO2) at 37°C. Cultures were passaged twice a week. hERG HEK-293 cells were cultured as described previously [41]. hERG transfected cells were maintained at 37°C in DMEM supplemented with 10% FCS, 1% penicillin-streptomycin, 2 mM l-glutamine, 0.1 mM non-essential amino acids, 1 mM sodium pyruvate, and 400 μg/ml geneticin (G-418). Cells were passaged weekly and were not allowed to become more than 80% confluent. For AFM studies, cells were seeded onto gelatin-coated glass slides, grown until 40–50% confluence. To perform TREC measurements, hERG cells were gently fixed with monomeric solution of glutaraldehyde [40].
Tip functionalization with hERG-extracellular antibody
AFM tips (standard Si3N4 tips (Veeco Instruments, USA) and magnetically coated tips (abbreviated as MAC tips) (Agilent Technologies, USA)) were extensively washed in chloroform and ethanol, then dried with nitrogen. Subsequently, to functionalize the tip surface (Si3N4) with amino (-NH2) (Fig. 2a) the tips were incubated in ethanolamine-HCl solution (Sigma, 550 mg/ml in dimethylsulfoxide (DMSO)) overnight [42]. Anti-KV11.1 (hERG-extracellular antibody) (Alomone Labs, Israel), which is directed against an extracellular epitope (corresponding to residues 430–445 of human Kv11.1) situated between S1 and S2 domains of the hERG subunits (Fig. 1) was then covalently linked to MAC tips via soft and long (∼8 nm) polyethylene glycol (PEG) as described previously [43]. Before the attachment of antibodies to the linker, the antibodies were purified from BSA molecules by high performance liquid chromatography (HPLC). As a linker, heterobifunctional PEG derivate, which contains aldehyde residue at one end and N-hydroxysuccinimide (NHS) on the other end, was used (Fig. 2). Tips were finally washed in Hank’s balanced salt solution (HBSS) containing 1.8 mM Ca2+ (AFM working buffer) and stored at 4°C.
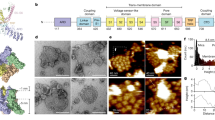
a Schematic representation of functionalization procedure of AFM tip with specific antibody via aldehyde-PEG-NHS cross-linker. b Scheme of dynamic recognition imaging to visualize hERG K+ channels (here binding sites an extracellular epitope between S1 and S2 domains of hERG subunit (shown in light grey)) on a gently fixed hERG HEK-293 cell surface
AFM recognition (TREC) imaging and force measurements
For the characterization of hERG HEK-293 cells grown on the glass slides, AFM images were taken in a contact mode configuration by using Si3N4 cantilevers (Veeco Instruments) with nominal spring constant of 10 pN/nm.
Recognition images were acquired in the magnetic AC (MAC) mode using a PicoPlus AFM (Agilent Technologies, USA) with MAC tips functionalized with anti-Kv11.1 (nominal spring constant of 100 pN/nm). All images were taken in HBSS containing 1.8 mM Ca2+ at room temperature with the same experimental conditions as previously discussed [40]. The TREC data were obtained by scanning ∼2×2 μm2 area of the cell surface with a lateral scan speed of ∼3.0 μm/s using a commercially available PicoTREC box (Agilent Technologies, USA). Briefly, PicoTREC records simultaneously two separate images by scanning the tethered sensor molecule across the surface to detect binding events. One image provides the information about the topography of the scanned surface. The second image displays a corresponding map of specifically recognized target molecules on the surface. To block the specific interactions between anti-Kv11.1 coated to the AFM tip and hERG HEK-293 cell surface, rErgtoxin-1 (ErgTx1; recombinant peptide, M.W.:4730, sequence: DRDSC VDKSR CAKYG YYQEC QDCCK NAGHN GGTCM FFKCK CA, Alomone Labs, Israel) (final concentration ∼1 μM) was gently injected into the fluid cell of the AFM during scanning.
For the evaluation of forces between anti-Kv11.1 and hERG HEK-293 cell surface in the absence and presence of rErgtoxin-1, standard AFM force spectroscopy was applied. In brief, this method requires functionalizing the AFM tip with anti-Kv11.1 antibody (see functionalization procedure above). After an antibody-functionalized tip contacts the cell surface, and a specific bond between the antibody and specific extracellular epitope (Fig. 1) can be formed. This bond will be ruptured (unbinding event) when the tip is pulled away from the cell surface. The amount by which the pulling cantilever bends before the bond ruptures is measured. From this value, the specific interaction (unbinding force) is calculated using the known spring constant, k, of the AFM cantilever. Such approach-retraction cycles (or force-distance cycles) were performed using anti-Kv11.1-coated cantilevers (rectangular cantilever, Veeco Instruments) with nominal spring constant of 20 pN/nm in the working buffer (HBSS, 1.8 mM Ca2+) at room temperature. The sweep-amplitude of the force-distance cycle was 2 μm at 1 Hz sweep rate. Force distance cycles were recorded on cell surfaces with the assistance of a CCD camera for positioning the AFM cantilever on cell patches formed by 3–5 cells (Fig. 3). More than 1,000 force-distance cycles were collected for each location on the surface of cells and up to four to five locations (different cells) for each condition (i.e. initial condition and addition of rErgtoxin-1). Usually one experiment was repeated three times and one typical experiment for each condition is shown in this study. Spring constants of cantilevers were determined using the thermal noise method [44] and analysis of interaction forces was performed using MATLAB Version 7 (MathWorks, Natick, MA, USA). To present the final force distributions empirical distribution density functions (or probability density functions (pdf)) were constructed as described previously [45].
Results
Morphology of hERG HEK-293 cells
To avoid significant changes in the hERG expression, hERG HEK-293 cells were grown with 40–50% confluence for further AFM investigations. Usually TREC and force measurements were conducted on the small cell patches, formed by three to five cells (Fig. 3). hERG HEK-293 show a similar morphology compared to normal HEK-293 cells. These cells are organized into typical triangular forms with ∼50–70 μm in length and heights varying from ∼200 nm at the periphery to ∼1–2 μm on the nucleus (Fig. 3).
Submicron localization of hERG K+ channels on hERG HEK-293 cells
The subcellular localization of hERG channels were achieved by immunofluorescence staining [35], which is a widely spread technique to visualize receptor binding sites on cell surfaces. It has been demonstrated that wild type hERG channels are distributed on the whole HEK-293 cellular surface [35]. In these optical studies, no information about topography is obtained and the observed lateral resolution is not better than 200 nm. At present, AFM, which represents a non-optical microscopy, offers a unique solution to obtain topography images with nanoscale resolution and single molecule interaction forces of biological specimens (e.g. proteins, DNA, membranes, cells, etc.) under/or near physiological conditions and without the need for rigorous sample preparation or labeling [46]. With the recent development of TREC (topography and recognition imaging), it becomes possible to quickly and easily obtain maps of binding sites with the lateral accuracy of several nanometers across variety surfaces, as it has been demonstrated on model receptor-ligand pairs [37, 39] and remodeled chromatin structures [38]. In addition, TREC represents an exquisite method to quickly obtain the local submicron distribution of receptors on cellular surface with an unprecedented lateral resolution of 5 nm [40].
In this study, TREC has been exploited to locally identify extracellular binding sites of hERG K+ channels on gently fixed hERG HEK-293 cells. Measurements were started with the scanning of whole cell surface with subsequent zooming into small areas ∼2×2 μm2. TREC images were acquired with magnetically coated AFM tips (MAC tips) that were functionalized with antibody anti-Kv11.1 (against epitope tags present on the hERG subunits (Fig. 2b)) via a soft and long (∼8 nm) polyethylene glycol (PEG)-linker (Fig. 2). All images were taken in Hank’s balanced salt solution (HBSS, 1.8 mM Ca2+) at 25°C. The oscillation amplitude was adjusted to be less than the extended PEG-linker to provide the proper recognition image with high efficiencies and repeatability (>90%) [38, 40]. Accordingly, the recognition map represents an amplitude reduction due to specific binding between anti-Kv11.1 on the tip and epitope tags on the cell surface (“dark” spots in Fig. 4a). These “dark” spots (amplitude reduction up to 2 nm) are distributed nonuniformly and reflect microdomains with dimensions from ∼30 up to ∼350 nm, with a mean ± SD of 99 ± 81 nm (n = 25) for the long domain axis. During several subsequent rescans recognition maps of hERG channels remain unchanged. Then, ErgTx1 was very slowly (∼50 μl/min) injected in the fluid cell while scanning the same sample. After the first and second injection of ErgTx1 (concentration of ∼400 nM), no visual changes in the recognition maps have been observed (Fig. 4 b). However, the recognition clusters partly disappeared after the third injection of ErgTx1 (∼1 μM) (Fig. 4c), whereas no change in the topography image has been observed (Fig. 4c′). The specific binding between anti-Kv11.1 and cellular surface was abolished when free ErgTx1 molecules bound to the hERG channels and thus blocked the antibody access to interact with epitope tags on hERG subunits. The topography of a scanned cell surface areas (Fig. 4a′, b′, and c′) shows a complex picture of linear and branched filamentous structures with some globular features. Most domains are found to be located near and between filaments (Fig. 5). TREC results suggest that ErgTx1 does not only interact with the extracellular surface of the pore domain (S5–S6), but might be to interact with the voltage sensing domains (S1–S4) of the hERG K+ channel.
Recognition images on a hERG HEK-293 cell surface. Simultaneous recorded recognition maps (a–c) and topography images (a′–c′) obtained with anti-Kv11.1—functionalized tip. Recognition events are depicted by threshold analysis (threshold = −1.7 nm) and the most pronounced recognition clusters are indicated by white lines. The presence of 400 nM ErgTx1 has not practically effect on recognition map (b), whereas the recognition clusters partly disappeared after the third injection of ErgTx1 (∼1 μM) (c). Blocking experiments do not affect membrane topography (compare a′, b′, and c′). Scale bars on all images are 170 nm
Recognition of hERG residues 430–445 with an AFM tip carrying epitope-specific antibody
In addition to the TREC measurements, AFM force-distance cycles with a tip carrying an epitope-specific antibody (anti-Kv11.1) were performed on living as well on gently fixed hERG HEK-293 cells as described previously [40]. Both studies on living and fixed cells conducted to the similar results (force distributions and binding probabilities), the data obtained with living cells are represented. The anti-Kv11.1 (HERG)-extracellular antibody is known to bind to the voltage sensor domain (S1–S2 region) of HERG K+ channel (Fig. 2). The specific binding of the antibody to the extracellular part of hERG channel was characterized by a unique unbinding force. To confirm the specificity of this binding, blocking experiments were carried out by injecting either free antibodies or free peptide antigen. In both cases, almost no unbinding events were observed. Binding probabilities (probability to record an unbinding event in force-distance cycles) from several experiments were as well quantified (Fig. 6a). The binding probability of ∼30% was calculated for the interaction between anti-Kv11.1-extracellular antibody and hERG HEK-293 cells. When free anti-Kv11.1 antibodies or free peptide antigens were present in solution, the binding probability drastically decreased to the level of ∼2% (Fig. 6a). By construction, an empirical probability density function (pdf) of the unbinding forces (Fig. 6b), the maximum of the distribution was found to be 45 ± 9 pN. As another indicator of the specificity, a very low binding probability (∼1.5%) with a force peak of ∼25 pN (Fig. 6) were found for the parent HEK-293 cells not expressing hERG K+ channels. These results illustrate that extracellular part of hERG K+ channel expressed in living cells can be specifically detected at the molecular level by using epitope-specific antibodies.
Recognition of hERG K+ channels on the surface of intact cells by an AFM tip carrying anti-Kv11.1. a Quantitative comparison of binding probabilities of anti-Kv11.1—coated tip on live hERG HEK-293 cells (left light grey) in the absence and presence of either free anti-Kv11.1 (middle light grey) or free antigen peptides (right light grey); binding probability of anti-Kv11.1—tip on parent HEK-293 cells is shown in black. The effect of ErgTx1 on the binding probabilities anti-Kv11.1—tip on hERG HEK-293 (grey). Values are mean ± SEM, n = 2,000–4,000. b Probability density functions (pdf) observed in the absence (solid lines) and in the presence of ErgTx1 (dot-dashed (300 nM) and dashed lines (1 μM)). Areas are scaled to the corresponding binding probabilities
We then investigated eventual effects on antibody binding induced by ErgTx1. Force curves were accumulated before and after ErgTx1 multiple injections at the same scan area with the same functionalized tip. In the presence of ErgTx1 at different concentrations, the peak force for force distributions (Fig. 6b) remains, whereas the binding probability between antibody and living hERG HEK 293 cells decreased dramatically after multiple ErgTx1 injections (Fig. 6a). These findings indicate additionally a possibly new binding site of ErgTx1 in the voltage sensor domain of hERG K+ channel.
Discussion
In this study, we focused on the microscopic mechanism by which ErgTx1 blocks K+ channels. There are two major classes of toxins, the pore-blocking [29, 47] and the gating-modifying [48, 49], which have been widely used to determine K+ channel structure and function. The interaction between toxins and the channel was studied by the two-electrode voltage-clamp recording techniques by many researchers. Typical pore-blocking toxins physically occlude the pore domain [29, 47] and bind to the outer pore domain [50]. The last is sensitive to changes in ion concentration [51] and to tetraethyl ammonium (known as external pore blocker) as well [52]. On the other hand, gating-modifier toxins bind to the voltage-sensor domain [48, 49]. It has been found that ErgTx1 binds to the outer vestibule of hERG [26, 27] like other pore blocking toxins and sensitive to tetraethyl ammonium [28]. However, the binging of ErgTx1 to the hERG are not affected by the changes in [K+] and in ionic strength of the extracellular solutions [27]. All these characteristics together with its incomplete blockade of hERG channel remind the behavior of BeKm-1 [23, 53]. To explain such mechanisms of toxin action, two hypothesis (“unconventional” gating modifiers [53] or only partial occlusion [54]) have been proposed and recently disproved in the case of ErgTx1 binding to hERG [55]. Furthermore, the most probable mechanism for incomplete block of hERG by ErgTx1 has been found to be a kinetic one [55].
In this study, we present data on a new binding site in the voltage sensor domain of ErgTx1 in the hERG K+ channel different than the pore binding site. To identify the possible binding sites of ErgTx1 on the voltage sensor domains of the hERG K+ channel, AFM measurements at the single-molecule level were conducted. We have demonstrated that AFM single-molecule recognition imaging (TREC) and spectroscopy are suitable methods to obtain information about the structure and function of hERG K+ channels on the surface of cells. Dynamic recognition imaging revealed non-uniform distribution of hERG K+ channels on the cell membrane at the submicron scale. In addition, to detect the presence of hERG K+ channels on the surface of living cells, interaction-force spectroscopy was performed. Both methods exploit AFM tips with very low surface density of ligands (∼400 molecules per μm2) to allow detection of single molecular events. Functionalization of AFM tips with anti-Kv11.1 (hERG)-extracellular antibody (Fig. 2a) enabled them to become monomolecular biosensors, which are capable of detecting binding sites of hERG (in S1–S2 domains) on the surface of cells expressed hERG channels. TREC was employed to identify anti-Kv11.1 (HERG)-extracellular antibody binding sites on gently fixed hERG HEK-293 cells and to co-localize their position with membrane topographical features. In addition, we have also investigated ErgTx1 blockade of the hERG K+ channel in a concentration dependent manner. The outcome of this study reveals that voltage sensing domain (S1-S4) of hERG K+ channel might be one of the binding sites of ErgTx1. The AFM methods used in this study illustrate a great potential for the investigation and localization of membrane proteins on the cell surface with several pN force resolution and a few nanometers positional accuracy.
References
Gurrola GB, Rosati B, Rocchetti M, Pimienta G, Zaza A, Arcangeli A, Olivotto M, Possani LD, Wanke E (1999) A toxin to nervous, cardiac, and endocrine ERG K+ channels isolated from Centruroides noxius scorpion venom. FASEB J 13:953–962
Sanguinetti MC, Jiang C, Curran ME, Keating MT (1995) A mechanistic link between an inherited and an acquired cardiac arrhythmia: HERG encodes the IKr potassium channel. Cell 81:299–307
Tseng GN (2001) IKr: the hERG channel. J Mol Cell Cardiol 33:835–849
Vandenberg JI, Walker BD, Campbell TJ (2001) HERG K+ channels: friend and foe. Trends Pharmacol Sci 22:240–246
Warmke JW, Ganetzky B (1994) A family of potassium channel genes related to eag in drosophila and mammals. Proc Natl Acad Sci USA 91:3438–3442
Trudeau MC, Warmke JW, Ganetzky B, Robertson GA (1995) HERG, a human inward rectifier in the voltage-gated potassium channel family. Science 269:92–95
Spector PS, Curran ME, Zou A, Keating MT, Sanguinetti MC (1996) Fast inactivation causes rectification of the IKr channel. J Gen Physiol 107:611–619
Wang S, Morales MJ, Liu S, Strauss HC, Rasmusson RL (1997) Modulation of HERG affinity for E-4031 by [K+]o and C-type inactivation. FEBS Lett 417:43–47
Rosati B, Marchetti P, Crociani O, Lecchi M, Lupi R, Arcangeli A, Olivotto M, Wanke E (2000) Glucose- and arginine-induced insulin secretion by human pancreatic ß-cells: the role of HERG K+ channels in firing and release. FASEB J 14:2601–2610
Smith GAM, Tsui H-W, Newell EW, Jiang X, Zhu X-P, Tsui FWL, Schlichter LC (2002) Functional up-regulation of HERG K+ channels in neoplastic hematopoietic cells. J Biol Chem 277:18528–18534
Roche O, Trube G, Zuegge J, Pflimlin P, Alanine A, Schneider G (2002) A virtual screening method for prediction of the hERG potassium channel liability of compound libraries. ChemBioChem 3:455–459
Mitcheson JS, Chen J, Sanguinetti MC (2000) Trapping of a methanesulfonanilide by closure of the HERG potassium channel activation gate. J Gen Physiol 115:229–239
Ho W-K, Kim I, Lee CO, Earm YE (1998) Voltage-dependent blockade of HERG channels expressed in Xenopus oocytes by external Ca2+ and Mg2+. J Physiol 507:631–638
Anumonwo JMB, Horta J, Delmar M, Taffet SM, Jalife J (1999) Proton and zinc effects on HERG currents. Biophys J 77:282–298
Po SS, Wang DW, Yang ICH, Johnson JP Jr, Nie L, Bennett PB (1999) Modulation of HERG potassium channels by extracellular magnesium and quinidine. J Cardiovasc Pharmacol 33:181–185
Johnson JP, Mullins Jr FM, Bennett PB (1999) Human ether-à-go-go–related gene K+ channel gating probed wit extracellular Ca2+. Evidence for two distinct voltage sensors. J Gen Physiol 113:565–580
Sanchez-Chapula JA, Sanguinetti MC (2000) Altered gating of HERG potassium channels by cobalt and lanthanum. Pflügers Arch 440:264–274
Ranganathan R, Lewis JH, MacKinnon R (1996) Spatial localization of the K+ channel selectivity filter by mutant cycle-based structure analysis. Neuron 16:131–139
Park CS, Miller C (1992) Mapping function to structure in a channel-blocking peptide: electrostatic mutants of charybdotoxin. Biochemistry 31:7749–7755
Tytgat J, Chandy KG, Garcia ML, Gutman GA, Martin- Eauclaire M-F, van der Walt JJ, Possani LD (1999) A unified nomenclature for short-chain peptides isolated from scorpion venoms: α-KTx molecular subfamilies. Trends Pharmacol Sci 20:444–447
Doyle DA, Morais Cabral J, Pfuetzner RA, Kuo A, Gulbis JM, Cohen SL, Chait BT, MacKinnon R (1998) The structure of the potassium channel: molecular basis of K+ conduction and selectivity. Science 280:69–77
Wang JM, Roh SH, Kim S, Lee CW, Kim JI, Swartz KJ (2004) Molecular surface of tarantula toxins interacting with voltage sensors in K(v) channels. J Gen Physiol 123:455–67
Korolkova YV, Kozlov SA, Lipkin AV, Pluzhnikov KA, Hadley JK, Filippov AK, Brown DA, Angelo K, Strøbæk D, Jespersen T, Olesen SP, Jensen BS, Grishin EV (2001) An ERG channel inhibitor from the Scorpion Buthus eupeus. J Biol Chem 276:9868–9876
Lecchi M, Redaelli E, Rosati B, Gurrola G, Florio T, Crociani O, Curia G, Restano Cassulini R, Masi A, Arcangeli A, Olivotto M, Schettini G, Possani LD, Wanke E (2002) Isolation of a long-lasting eag-related gene-type K+ current in MMQ lactrotrophs and its accommodating role during slow firing and prolactin release. J Neurosci 22:3414–3425
Nastainczyk W, Meves H, Watt DD (2002) A short-chain peptide toxin isolated from Centruroides sculpturatus scorpion venom inhibits ether-à-go-go-related gene K+ channels. Toxicon 40:1053–1058
Pardo-Lopez L, Garcia-Valdes L, Gurrola GB, Robertson GA, Possani LD (2002) Mapping the receptor site for ergtoxin, a specific blocker of ERG channels. FEBS Lett 510:45–49
Pardo-Lopez L, Zhang M, Liu J, Jiang M, Possani LD, Tseng GN (2002) Mapping the binding site of a human ether-a-go-go related gene-specific peptide toxin (ErgTx) to the channel’s outer vestibule. J Biol Chem 277:16403–16411
Torres AM, Bansal P, Alewood PF, Bursill JA, Kuchel PW, Vandenberg JI (2003) Solution structure of CnErg1 (Ergtoxin), a HERG specific scorpion toxin. FEBS Lett 539:138–142
Rodriguez de la Vega RC, Merino E, Becerril B, Possani LD (2003) Novel interactions between K+ channels and scorpion toxins. Trends Pharmacol Sci 24:222–227
Xu CQ, Zhu SY, Chi CW, Tytgat J (2003) Turret and pore block of K+ channels: what is the difference? Trends Pharmacol Sci 24:446–449
Clarke CE, Hill AP, Zhao J, Kondo M, Subbiah RN, Campbell TJ, Vandenberg JI (2006) Effect of S5P alpha-helix charge mutants on inactivation of hERG K+ channels. J Physiol 573:291–304
Pfister G, Stroh CM, Pershinka H, Kind M, Knoflach M, Hinterdorfer P, Wick G (2005) Detection of HSP60 on the membrane surface of stressed human endothelial cells by atomic force and confocal microscopy. J Cell Sci 118:1587–1594
Puntheeranurak T, Wildling L, Gruber HJ, Kinne RKH, Hinterdorfer P (2006) Ligands on the string: single-molecule AFM studies on the interaction of antibodies and substrates with Na+-glucose co-transporter SGLT1 in living cells. J Cell Sci 119:2960–2967
Hinterdorfer P, Dufrêne YF (2006) Detection and localization of single molecular recognition events using atomic force microscopy. Nat Methods 3:347–355
Zhou Z, Gong Q, Epstein ML, January CT (1998) HERG channel dysfunction in human long QT syndrome. J Biol Chem 273:21061–21066
Schütz GJ, Pastushenko VPh, Gruber HJ, Knaus H-G, Pragl B, Schindler H (2000) 3D imaging of individual ion channels in live cells at 40 nm resolution. Single Mol 1:25–31
Stroh CM, Ebner A, Geretschläger M, Freudenthaler G, Kienberger F, Kamruzzahan ASM, Smith-Gill SJ, Gruber HJ, Hinterdorfer P (2004) Simultaneous topography and recognition imaging using force microscopy. Biophys J 87:1981–1990
Stroh C, Wang H, Bash R, Ashcroft B, Nelson J, Gruber H, Lohr D, Lindsay SM, Hinterdorfer P (2004) Single-molecule recognition imaging microscopy. Proc Natl Acad Sci USA 101:12503–12507
Ebner A, Kienberger F, Kada G, Stroh CM, Geretschläger M, Kamruzzahan ASM, Wildling L, Johnson WT, Ashcroft B, Nelson J, Lindsay SM, Gruber HJ, Hinterdorfer P (2005) Localization of single avidin-biotin interactions using simultaneous topography and molecular recognition imaging. ChemPhysChem 6:897–900
Chtcheglova LA, Waschke J, Wildling L, Drenckhahn D, Hinterdorfer P (2007) Nano-scale dynamic recognition imaging on vascular endothelial cells. Biophys J 93:L11–L13
Zhou Z, Gong Q, Ye B, Fan Z, Makielevski JC, Robertson CT (2006) Properties of HERG channels stably expressed in HEK 293 cells studied in physiological temperature. Biophys J 74:230–241, January
Riener CK, Stroh CM, Ebner A, Klampfl C, Gall AA, Romanin C, Lyubchenko YL, Hinterdorfer P, Gruber HJ (2003) Simple test for single molecule recognition force microscopy. Anal Chim Acta 479:59–75
Ebner A, Wildling L, Kamruzzahan ASM, Rankl C, Wruss J, Hahn CD, Hölz M, Zhu R, Kienberger F, Blaas D, Hinterdorfer P, Gruber HJ (2007) A new, simple method for linking of antibodies to atomic force microscopy tips. Bioconjugate Chem 18:1176–1184
Hutter J, Bechhoefer J (1993) Calibration of atomic-force microscope tips. Rev Sci Inst 64:1868–1873
Baumgartner W, Hinterdorfer P, Schindler H (2000) Data analysis of interaction forces measured with the atomic force microscope. Ultramicroscopy 82:85–95
Hörber JKH, Miles MJ (2003) Scanning probe evolution in biology. Science 302:1002–1005
Garcia ML, Gao Y, McManus OB, Kaczorowski GJ (2001) Potassium channels: from scorpion venoms to high-resolution structure. Toxicon 39:739–748
Swartz KJ, MacKinnon R (1997) Mapping the receptor site for hanatoxin, a gating modifier of voltage-dependent K+ channels. Neuron 18:675–682
Wang J, Salata JJ, Bennett PB (2003) Saxitoxin is a gating modifier of hERG K+ channels. J Gen Physiol 121:583–598
MacKinnon R, Heginbotham L, Abramson T (1990) Mapping the receptor site for charybdotoxin, a pore-blocking potassium channel inhibitor. Neuron 5:767–771
Goldstein SA, Miller C (1993) Mechanism of charybdotoxin block of a voltage-gated K+ channel. Biophys J 65:1613–1619
Miller C (1988) Competition for block of a Ca2+-activated K+ channel by charybdotoxin and tetraethylammonium. Neuron 1:1003–1006
Zhang M, Korolkova YV, Liu J, Tiang M, Grishin EV, Tseng GN (2003) BeKm-1 is a hERG-specific toxin that shares the structure with ChTx but the mechanism of action with ErgTx1. Biophys J 84:3022–3036
Korolkova YV, Tseng GN, Grishin EV (2004) Unique interaction of scorpion toxins with the hERG channel. J Mol Recognit 17:209–217
Hill AP, Sunde M, Campbel TJ, Vandenberg JI (2007) Mechanism of block of the hERG K+ channel by the scorpion toxin CnErg1. Biophys J 92:3915–3929
Acknowledgement
We thank Dr. Hermann Gruber and Prof. Vasilii Pastushenko for the enlightening discussions. This work was supported in part by grants from the Istanbul University, BAP (project no: 149/20082003 to FA) and European Commission (contract no. LSHGCT-2005–512101 to LW and “Bio-Light-Touch” LAC and PH). Chtcheglova and Atalar have equally contributed to this work.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
About this article
Cite this article
Chtcheglova, L.A., Atalar, F., Ozbek, U. et al. Localization of the ergtoxin-1 receptors on the voltage sensing domain of hERG K+ channel by AFM recognition imaging. Pflugers Arch - Eur J Physiol 456, 247–254 (2008). https://doi.org/10.1007/s00424-007-0418-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00424-007-0418-9