Abstract
Purpose
Infections are common complications in patients following liver transplantation (LTX). The early diagnosis and prognosis of these infections is an unmet medical need even when using routine biomarkers such as C-reactive protein (CRP) and procalcitonin (PCT). Therefore, new approaches are necessary.
Methods
In a prospective, observational pilot study, we monitored 30 consecutive patients daily between days 0 and 13 following LTX using the 29-mRNA host classifier IMX-BVN-3b that determine the likelihood of bacterial infections and viral infections. True infection status was determined using clinical adjudication. Results were compared to the accuracy of CRP and PCT for patients with and without bacterial infection due to clinical adjudication.
Results
Clinical adjudication confirmed bacterial infections in 10 and fungal infections in 2 patients. 20 patients stayed non-infected until day 13 post-LTX. IMX-BVN-3b bacterial scores were increased directly following LTX and decreased until day four in all patients. Bacterial IMX-BVN-3b scores detected bacterial infections in 9 out of 10 patients. PCT concentrations did not differ between patients with or without bacterial, whereas CRP was elevated in all patients with significantly higher levels in patients with bacterial infections.
Conclusion
The 29-mRNA host classifier IMX-BVN-3b identified bacterial infections in post-LTX patients and did so earlier than routine biomarkers. While our pilot study holds promise future studies will determine whether these classifiers may help to identify post-LTX infections earlier and improve patient management.
Clinical trial notation
German Clinical Trials Register: DRKS00023236, Registered 07 October 2020, https://drks.de/search/en/trial/DRKS00023236
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In patients suffering from end-stage liver disease (ESLD), liver transplantation (LTX) is a well-established curative therapeutic option [1, 2]. The outcomes of these patients have significantly improved by applying optimized posttransplant care. A key complication in these patients are severe infections, which may lead to transplant failure and/or increased mortality [3]. Most infections are of bacterial etiology and occur within the first four weeks following LTX [4, 5]. Routinely used biomarkers (such as C-reactive protein (CRP) and procalcitonin (PCT)) have limited specificity and sensitivity [6,7,8]. Direct detection methods, such as microbiological cultures and/or molecular methods, can be insensitive [9,10,11]. Therefore, new diagnostics are needed to allow for early and accurate detection of infections and initiation of early antimicrobial therapy.
The transcriptomic classifier IMX-BVN-3b has been reported to allow early detection of bacterial and viral infections [12]. IMX-BVN-3b is based on the expression of 29 host-response messenger RNA (mRNA) and is interpreted by a machine-learning algorithm [13]. The accuracy of this classifiers has been demonstrated in critically ill surgical and medical ICU patients treated in emergency departments [12, 14,15,16]. The accuracy for the diagnosis and prognosis of infections in transplant patients has not yet been determined.
In the present study we investigated the accuracy of the IMX-BVN-3bclassifiers to diagnose and prognose infections in post-LTX patients in comparison to routinely used biomarkers for the detection of bacterial and viral infections. Moreover, the kinetics of classifier results were determined between day 0 and 13 post-LTX.
Materials and methods
Study design
The observational single-center pilot study was approved by the local ethics committee (Ethics Committee of the Medical Faculty of Heidelberg, Trial Code No. S-693/2020 / German Clinical Trials Register: DRKS00023236) and conducted in the surgical unit of the Heidelberg University Hospital, Germany between July 2021 and February 2023. All study participants gave written informed consent. In total, 30 patients suffering from ESLD undergoing orthotopic LTX were enrolled in this study, representing the only inclusion criterion. Missing written consent and age below 18 years were defined as exclusion criteria. The treatment of patients was based on the Heidelberg Manual for LTX [17]. Blood was collected in PAXgene® Blood RNA tubes (PreAnalytix, Hombrechtikon, Switzerland) daily between days 0 and 13 post-LTX for testing of IMX-BVN-3b and IMX-SEV-3b classifiers; plasma samples were collected at the same time points post-LTX to determine CRP and PCT concentrations. Relevant baseline data (demographic data, primary site of infection), clinical data (disease severity scores, such as Simplified Acute Physiology (SAPS II)-score, Sequential Organ Failure Assessment (SOFA)-score and Acute Physiology Health Evaluation (APACHE II)-score, surgical procedures, antifungal therapy, outcome parameters) and routine infection parameters (leukocytes, C-reactive protein (CRP), procalcitonin (PCT), body temperature), according to the standardized central laboratory protocols, were also collected.
The primary endpoint of this study was the accuracy of the IMX-BVN-3b for the detection of bacterial and viral infections following LTX as determined by clinical adjudication. Secondary endpoints included a) the kinetics of IMX-BVN-3b post-LTX and b) the comparison of IMX-BVN-3b accuracy to biomarkers CRP and PCT.
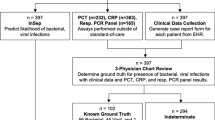
A detailed study flowchart is shown in Fig. 1. The study was performed according to the Strobe statement (Supplementary Material 1).
Clinical adjudication
The reference standard was determined by a chart review to establish the true infection status for the presence or absence of a bacterial, viral and/or other infection. The expert panel consisted of two physicians trained in infectious diseases who classified the patients for bacterial, viral and fungal infections according to previously published criteria[18, 19] and in accordance to Centers for Disease Control and Prevention (CDC) /National Healthcare Safety Network (NHSN) criteria for bacterial infections as well as European Organization for Research and Treatment of Cancer (EORTC) criteria for invasive fungal infections [20, 21]. They used medical history, physical examination findings, all available laboratory, including CRP and PCT, microbiological and imaging data, and the discharge report or treatment data. The expert panel was blinded to the IMX-BVN-3b results.
Blood sample collection in PAXgene tube and amplification of target genes
2.5 ml of whole blood were obtained daily using the PAXgene® Blood RNA tube. The tubes were frozen at -80 °C. Pseudonymized samples were shipped in batches to the Inflammatix laboratory (Sunnyvale, CA, USA) for gene amplification and IMX-BVN-3b classifier processing as previously described [12]. Operators at Inflammatix were blinded to any other study results.
IMX-BVN-3b classifiers
The machine learning algorithms IMX-BVN-3b read and interpreted results of amplification for the 29 host mRNAs [22]. Results of the IMX-BVN-3b classifier were provided as numerical scores that fall into one of five interpretation bands for the likelihood of a bacterial infection, and into one of five interpretation bands for the likelihood of a viral infection. Predefined thresholds are used to categorize results into the Very low, Low, Moderate, High, or Very high bacterial and viral interpretation bands. Classifier results were provided to the Heidelberg study team for statistical analysis.
Statistical analysis
All data were entered into an electronic database (Excel 2021; Microsoft Corp, Redmond, USA) and analyzed using SPSS software (Version 28.0; SPSS, Inc., Chicago, USA). Figures were drawn with GraphPad Prism 10 (GraphPad Software, La Jolla, USA), SPSS software and assembled with the presentation software PowerPoint 2021 (Microsoft Corp, Redmond, USA). Categorical data were shown as absolute and relative frequencies. Quantitative data were presented as median with quartiles. The Kolmogorov–Smirnov test was used to check for normal distribution. Due to non-normally distributed data, non-parametric methods for evaluation were used (Chi-square test for categorical data, Mann–Whitney U test for continuous data). Receiver-Operating characteristics were used to assess discrimination. Spearman correlation was used to detect significant correlations between different scores. We introduced virtual timepoints (V) for the first bacterial infection to normalize the analysis. Furthermore, infected patients were compared to a comparison group including non-infected age- and sex-matched patients. Moreover patients were classified in bands according to predefined cut off values, regarding IMX-BVN-3b [12, 14], PCT [23, 24] and CRP [25,26,27] for prediction of bacterial infections and calculation of likelihood ratios. A p-value < 0.05 was considered statistically significant.
Results
Patient characteristics and microbiological findings
Between day 0 to day 13 post LTX, 10 (33.3%) of 30 patients developed a bacterial infection as determined by clinical adjudication whereas 20 patients showed no signs of bacterial infections; there were no viral infections during the observation period. Anatomical sites of bacterial infection were intraabdominal (n = 6), respiratory (pneumonia, n = 1) and urinary tract (n = 1); two patients developed bacteriemia with positive blood cultures. Invasive fungal infections were detected in two (6.7%) patients; one with Aspergillus fumigatus in the lung and one with Candida albicans in the abdomen, 6 (20%) patients were colonized with fungal pathogens (Candida species). General clinical characteristics of patients are presented in Table 1, clinical characteristics of ICU and hospital stay are shown in Table 2. Patients with or without bacterial infection did not differ significantly with regard to underlying diseases, immunosuppressive medications or other clinical characteristics (Table 1). Patients with bacterial infections had significantly longer hospital and ICU length of stay; they also showed more significant blood loss during surgery and a significantly higher proportion of patients with bacterial infections received platelets (Table 2).
IMX-BVN-3b for the diagnosis of bacterial infections
IMX-BVN-3b scores were at high scores in all patients directly following LTX (Fig. 2a). In patients with bacterial infections, scores were significantly higher and stayed high compared to noninfected patients (Supplementary Fig. 1), in whom scores decreased over time. In contrast, the routinely used infection parameters PCT, CRP and white blood cell count showed limited value in the detection of bacterial infections (Fig. 2b-d and Supplementary Figs. 2a-c). PCT levels were highly increased starting from day one following LTX in all patients and declined in the days after, independently from the occurrence of bacterial infections (Fig. 2b and Supplementary Fig. 2a). CRP levels were increased in all patients starting from day one following LTX. In uninfected patients CRP declined in the days afterwards, whereas in patients with proven bacterial infection CRP showed significant elevated levels. Nevertheless, all patients presented with values indicating an inflammatory reaction using the cut-offs for uninfected patients (Fig. 2c and Supplementary Fig. 2b). White blood cell count showed undulating values in all patients with a trend to lower values in patients without bacterial infections compared to patients with bacterial infections without showing relevant significant differences (Fig. 2d and Supplementary Fig. 2c).
Bacterial IMX-BVN-3b scores and inflammatory markers in patients following liver transplantation. (a) Bacterial IMX-BVN-3b scores were measured in patients following liver transplantation. Patients were divided according to the occurrence of a proven bacterial infection within the first 14 days following LTX. Bacterial IMX-BVN-3b scores are presented for unobtrusive patients as green line combined with light grey area and for patients with bacterial infections as red line combined with a dark grey area. Plasma samples were collected immediately following liver transplantation (d0), and the days afterwards until day 13 as indicated. Data are presented as median with 95% Confidence interval as borders of the areas. (b) Procalcitonin (PCT) was measured in patients following liver transplantation. Patients were divided according to the occurrence of a proven bacterial infection within the first 14 days following LTX. PCT levels are presented for unobtrusive patients as green line combined with light grey area and for patients with bacterial infections as red line combined with a dark grey area. Plasma samples were collected immediately following liver transplantation (d0), and the days afterwards until day 13 as indicated. Data are presented as median with 95% Confidence interval as borders of the areas. (c) C-reactive protein (CRP) was measured in patients following liver transplantation. Patients were divided according to the occurrence of a proven bacterial infection within the first 14 days following LTX. CRP levels are presented for unobtrusive patients as green line combined with light grey area and for patients with bacterial infections as red line combined with a dark grey area. Plasma samples were collected immediately following liver transplantation (d0), and the days afterwards until day 13 as indicated. Data are presented as median with 95% Confidence interval as borders of the areas. (d) White blood cell counts were measured in patients following liver transplantation. Patients were divided according to the occurrence of a proven bacterial infection within the first 14 days following LTX. White blood cell counts are presented for unobtrusive patients as green line combined with light grey area and for patients with bacterial infections as red line combined with a dark grey area. Plasma samples were collected immediately following liver transplantation (d0), and the days afterwards until day 13 as indicated. Data are presented as median with 95% Confidence interval as borders of the areas
These findings could be proven within the likelihood analysis of IMX-BVN-3b, CRP and PCT (Supplementary Table 1 A to C), indicating the best performance for IMX-BVN-3b around day 6 to 9 following LTX, correlating with the fact that most bacterial infections occurred during this period.
In a subsequent analysis, focusing on the timepoint of the bacterial infection, IMX-BVN-3b was also able to correctly identify patients with bacterial infections at least one day before the clinical diagnosis (Fig. 3a and Supplementary Fig. 3a), which could be verified by a ROC-analysis (Supplementary Fig. 3b). In contrast, PCT was unable to significantly differentiate between patients with and without bacterial infections (Fig. 3b). On the other hand, CRP showed a comparable course to IMX-BVN-3b for the diagnosis of bacterial infection, when focusing on the timepoint of infection (Fig. 3c). Regarding the white blood cell count, no significant differences could be observed in patients with or without bacterial infection at timepoint of the clinical diagnosis (Fig. 3d). These findings could be proven within the likelihood analysis of IMX-BVN-3b, CRP and PCT (Table 3 A to C). Moreover, in detailed analysis of IMX-BVN-3b curves of each single patient, nine of the 10 patients with adjudicated bacterial infection were correctly identified by the test (Supplementary Fig. 3a).
Bacterial IMX-BVN-3b scores and inflammatory markers in patients following liver transplantation adjusted to the timepoint of the bacterial infection. (a) Bacterial IMX-BVN-3b scores were measured in patients following liver transplantation. Measurements in patients with bacterial infection were adjusted to the timepoint of appearance (grey box) and then compared to non-infected patients (white box). In patients with a bacterial infection, new timepoints were created by matching them to the first time of bacterial infection, whereas the control group without bacterial infection was created by matching them in an age- and sex-related manner to the same timepoints of patients with a bacterial infection. The following virtual timepoints were created: the first measurement two days before the first bacterial infection (preV-2), the first measurement two days before the first bacterial infection (preV-1), the plasma level at the time of bacterial infection (V0) and the next measured plasma levels on the days afterwards (V1-V4). Data in box plots are given as median, 25th percentile, and 75th percentile, with the 10th and 90th percentile at the end of the whiskers. Concerning symbolism and higher orders of significance: p < 0.05: *; p < 0.01: **; p > 0.001: ***Mann–Whitney U Test. (b) Procalcitonin (PCT) was measured in patients following liver transplantation. Measurements in patients with bacterial infection were adjusted to the timepoint of appearance (grey box) and then compared to non-infected patients (white box). In patients with a bacterial infection, new timepoints were created by matching them to the first time of bacterial infection, whereas the control group without bacterial infection was created by matching them in an age- and sex-related manner to the same timepoints of patients with a bacterial infection. The following virtual timepoints were created: the first measurement two days before the first bacterial infection (preV-2), the first measurement two days before the first bacterial infection (preV-1), the plasma level at the time of bacterial infection (V0) and the next measured plasma levels on the days afterwards (V1-V4). Data in box plots are given as median, 25th percentile, and 75th percentile, with the 10th and 90th percentile at the end of the whiskers. Concerning symbolism and higher orders of significance: p < 0.05: *; p < 0.01: **; p > 0.001: ***Mann–Whitney U Test. (c) C-reactive protein (CRP) was measured in patients following liver transplantation. Measurements in patients with bacterial infection were adjusted to the timepoint of appearance (grey box) and then compared to non-infected patients (white box). In patients with a bacterial infection, new timepoints were created by matching them to the first time of bacterial infection, whereas the control group without bacterial infection was created by matching them in an age- and sex-related manner to the same timepoints of patients with a bacterial infection. The following virtual timepoints were created: the first measurement two days before the first bacterial infection (preV-2), the first measurement two days before the first bacterial infection (preV-1), the plasma level at the time of bacterial infection (V0) and the next measured plasma levels on the days afterwards (V1-V4). Data in box plots are given as median, 25th percentile, and 75th percentile, with the 10th and 90th percentile at the end of the whiskers. Concerning symbolism and higher orders of significance: p < 0.05: *; p < 0.01: **; p > 0.001: ***Mann–Whitney U Test. (d) White blood cell counts were measured in patients following liver transplantation. Measurements in patients with bacterial infection were adjusted to the timepoint of appearance (grey box) and then compared to non-infected patients (white box). In patients with a bacterial infection, new timepoints were created by matching them to the first time of bacterial infection, whereas the control group without bacterial infection was created by matching them in an age- and sex-related manner to the same timepoints of patients with a bacterial infection. The following virtual timepoints were created: the first measurement two days before the first bacterial infection (preV-2), the first measurement two days before the first bacterial infection (preV-1), the plasma level at the time of bacterial infection (V0) and the next measured plasma levels on the days afterwards (V1-V4). Data in box plots are given as median, 25th percentile, and 75th percentile, with the 10th and 90th percentile at the end of the whiskers. Concerning symbolism and higher orders of significance: p < 0.05: *; p < 0.01: **; p > 0.001: ***Mann–Whitney U Test
IMX-BVN-3b for the detection of viral infections following LTX
Clinical adjudication did not diagnose any viral infections. Viral IMX-BVN-3b scores did not increase during the observation period (Supplementary Fig. 4).
IMX-BVN-3b in patients with fungal infections vs. fungal colonization
Clinical adjudication confirmed invasive fungal infections in two patients, whereas 6 patients were found to be colonized with fungi. IMX-BVN-3b bacterial and viral scores did not increase in patients with fungal infections. Bacterial and viral IMX-BVN-3b scores were similar in patients with fungal colonization compared to the uninfected patients (data not shown).
Discussion
The present study shows the potential utility of the IMX-BVN-3b test for the earlier detection of bacterial infections following LTX and differentiation of uninfected patients.
Bacterial infections following LTX are common events due to the need of immunosuppression and surgical procedures, especially in the early period of post-transplant care [4, 5]. The routinely used culture diagnostics have long turnaround times until results are available and are associated with relevant weakness, due to false positive or negative results [28, 29]. Available molecular diagnostics approaches including polymerase-chain reaction (PCR) analysis, which is linked with limited number of detected pathogens or next-generation-sequencing methods have been tested in several study settings [19, 30], but are far away from being used as routine methods.
Plasma biomarkers, like CRP, PCT or white blood cell count are routinely used in daily clinical care but have relevant limitations [11, 31]. Therefore, new diagnostic approaches are needed, as time delays in the reliable diagnostic of bacterial infections in patients following LTX might result in increased morbidity and mortality of the affected patient [3].
The combined host response signature test IMX-BVN-3b is still under development by Inflammatix, Inc. The test is able to quantify 29 messenger RNA (mRNA) expressed in whole blood, which were derived from transcriptomic studies in patients with critical illness or sepsis [32,33,34]. Using a machine learning classifier, the prognostic performance in patients with sepsis or severe illness was optimized [35]. Nevertheless, this is also a limiting point, as the test has not been solely examined in patients following liver transplantation or other solid organ transplantation until now. Also, the test is not yet commercially available. Despite these limitations, already published data showed promising results of earlier, reliable diagnosis in critical ill patients [12, 16]. Patients following LTX are good comparators to patients with severe sepsis and maybe more in danger due to the need of immunosuppressive drugs to avoid rejection, which increases the risk of severe infections, with an inadequate response of the immune system, leading to a severe critical illness [4, 5]. As already described above, routinely used diagnostics methods, e.g., CRP or PCT are fraught with limitations, which has also been shown in our department [36]. Moreover, PCT levels were described to be increased directly after liver transplantation without a relevant source of infection, presumably due to a transfer from the donor [37, 38], rendering it in particular as a single parameter to be less usable in patients following LTX [39]. Therefore, the test that may be commercially available in the future that includes the IMX-BVN-3b classifier (or a future version of it) might be a promising new rapid (~ 30 min) diagnostic option [40]. As shown here in the presented study, bacterial IMX-BVN-3b scores are highly elevated in all patients directly following LTX, potentially signaling a severe bacterial infection, which might be due to a donor-received factor introduced during the transplantation from the donor organ, comparable to PCT. In contrast to PCT, bacterial IMX-BVN-3b scores dropped to low scores in patients without proven bacterial infection, whereas scores in patients with infections stayed high, indicative of the existing inflammation. Therefore, bacterial IMX-BVN-3b may be an additional option in routine care to optimize the diagnostic options in patients following LTX with comparable performance as shown in patients with sepsis [12, 15]. This point is highlighted by the fact, that adjusted to the timepoint of bacterial infection, bacterial IMX-BVN-3b showed high scores one day before the clinical diagnosis of the bacterial infections comparable to CRP.
Viral infections, like cytomegalovirus, in patients following LTX may also lead to increased morbidity and poor outcome of the affected patient as the virus can induce acute rejections [41, 42]. Most of the viral infections are due to a virus reactivation, despite antiviral prophylaxis, following first virus contact and long before transplantation due to persistence within the body which may lead to a delayed diagnosis [43,44,45]. Therefore, a fast and reliable diagnosis of such infections is necessary. In the present study, none of the patients showed signs of a viral infection. These findings were also proven by the viral IMX-BVN-3b score. Within the whole cohort, viral IMX-BVN-3b levels stayed at low levels, indicating no signs of a probable viral infection. Nevertheless, these findings should be reevaluated in a larger cohort, that includes patients exhibiting a viral infection post-LTX.
The fact, that IMX-BVN-3b was not able to differ between patients with fungal infection and without such an infection might be explained due to the underlying data base of the test, which was not developed for fungal infections. As this is a machine learning system [22], further development approaches might offer the option to detect fungal infections in patients following LTX as well as suffering by other diseases.
Limitations
This study had several limitations. Due to the ongoing Covid-19 pandemic in 2020 and 2021 the planned number of 50 patients could not be reached as the rates of transplantation were reduced. Due to organizational and feasibility reasons, the recruitment was stopped after 30 patients. All planned laboratory measurements were carried out and revealed plausible results. Moreover, the observation period was limited to just the first 14 days following LTX which was combined with a long-term observation period to prove the outcome of each single patient. Therefore, infections at later timepoints than 14 days may have been missed. Enlarging the observation period could have led to an increased burden for each single patient due to the need for more blood samples as well as for the study teams to ensure the feasibility of the study. Nevertheless, the presented results are conclusive, but will need to be verified within a larger study in the future.
Conclusions
This study showed for the first time the kinetics of the 29-mRNA classifier IMX-BVN-3b in the early post-surgery care of patients following LTX and offered promising results for the earlier detection of bacterial infections in this time period. Moreover, it may have additional value on the routinely used inflammatory markers (CRP, PCT) and microbiological diagnostics for treatment decision, especially in cases of difficult conditions and uncertainty of a need for treatment. The viral score of the 29-mRNA classifier was able to rule out viral infections following LTX.
Data availability
The data presented in this study are available on request from the corresponding authors.
Abbreviations
- APACHE II:
-
Acute Physiology Health Evaluation score
- ARF:
-
Acute renal failure
- BDA:
-
Biliodigestive anastomosis
- BMI:
-
Body Mass Index
- CDC:
-
Centers for Disease Control and Prevention
- CKD:
-
Chronic kidney disease
- CRP:
-
C-reactive protein
- EORTC:
-
European Organisation for Research and Treatment of Cancer
- FFP:
-
Fresh frozen plasma
- HCC:
-
Hepatocellular carcinoma,
- ICU:
-
Intensive care unit
- LTX:
-
Liver transplantation
- MELD:
-
Model of end-stage liver disease
- mRNA:
-
Messenger RNA
- n.a:
-
Not applicable
- NASH:
-
Non-alcoholic Fatty Liver Disease
- NHSN:
-
National Healthcare Safety Network
- n.o.p.:
-
Number of patients
- NYHA:
-
New York Heart Association
- PBC:
-
Primary Biliary Cirrhosis
- PCR:
-
Polymerase-chain reaction
- PCT:
-
Procalcitonin
- PRBC:
-
Packed red blood cells
- PSC:
-
Primary sclerosing cholangitis
- SAPS II:
-
Simplified Acute Physiology score,
- SDC:
-
Supplemental Digital Content
- SOFA:
-
Sequential Organ Failure Assessment score
- V:
-
Virtual timepoint
References
Trunecka P (2013) Immunosuppression after liver transplant, now and in future. Vnitr Lek 59:671–677
Lucey MR, Furuya KN, Foley DP (2023) Liver Transplantation. N Engl J Med 389:1888–1900. https://doi.org/10.1056/NEJMra2200923
Winston DJ, Emmanouilides C, Busuttil RW (1995) Infections in liver transplant recipients. Clin Infect Dis 21:1077–1089
Fishman JA (2007) Infection in solid-organ transplant recipients. N Engl J Med 357:2601–2614. https://doi.org/10.1056/NEJMra064928
Kim SI (2014) Bacterial infection after liver transplantation. World J Gastroenterol 20:6211–6220. https://doi.org/10.3748/wjg.v20.i20.6211
Christ-Crain M, Muller B (2005) Procalcitonin in bacterial infections–hype, hope, more or less? Swiss Med Wkly 135:451–460. https://doi.org/10.4414/smw.2005.11169
de Oliveira VM, Moraes RB, Stein AT, Wendland EM (2017) Accuracy of C - Reactive protein as a bacterial infection marker in critically immunosuppressed patients: A systematic review and meta-analysis. J Crit Care 42:129–137. https://doi.org/10.1016/j.jcrc.2017.07.025
Svaldi M, Hirber J, Lanthaler AI, Mayr O, Faes S, Peer E, Mitterer M (2001) Procalcitonin-reduced sensitivity and specificity in heavily leucopenic and immunosuppressed patients. Br J Haematol 115:53–57. https://doi.org/10.1046/j.1365-2141.2001.03083.x
Biron BM, Ayala A, Lomas-Neira JL (2015) Biomarkers for Sepsis: What Is and What Might Be? Biomark Insights 10:7–17. https://doi.org/10.4137/BMI.S29519
Gur A, Oguzturk H, Kose A, Turtay MG, Ersan V, Bayindir Y, Ince V, Gurbuz S, Yucel N (2017) Prognostic Value of Procalcitonin, CRP, Serum Amyloid A, Lactate and IL-6 Markers in Liver Transplant Patients Admitted to ED with Suspected Infection. In Vivo 31:1179–1185. https://doi.org/10.21873/invivo.11187
Zant R, Melter M, Knoppke B, Ameres M, Kunkel J (2014) Kinetics of interleukin-6, procalcitonin, and C-reactive protein after pediatric liver transplantation. Transplant Proc 46:3507–3510. https://doi.org/10.1016/j.transproceed.2014.08.048
Bauer W, Kappert K, Galtung N, Lehmann D, Wacker J, Cheng HK, Liesenfeld O, Buturovic L, Luethy R, Sweeney TE et al (2021) A Novel 29-Messenger RNA Host-Response Assay From Whole Blood Accurately Identifies Bacterial and Viral Infections in Patients Presenting to the Emergency Department With Suspected Infections: A Prospective Observational Study. Crit Care Med 49:1664–1673. https://doi.org/10.1097/CCM.0000000000005119
Mayhew MB, Buturovic L, Luethy R, Midic U, Moore AR, Roque JA, Shaller BD, Asuni T, Rawling D, Remmel M et al (2020) A generalizable 29-mRNA neural-network classifier for acute bacterial and viral infections. Nat Commun 11:1177. https://doi.org/10.1038/s41467-020-14975-w
Brakenridge SC, Chen UI, Loftus T, Ungaro R, Dirain M, Kerr A, Zhong L, Bacher R, Starostik P, Ghita G et al (2022) Evaluation of a Multivalent Transcriptomic Metric for Diagnosing Surgical Sepsis and Estimating Mortality Among Critically Ill Patients. JAMA Netw Open 5:e2221520. https://doi.org/10.1001/jamanetworkopen.2022.21520
Brakenridge SC, Starostik P, Ghita G, Midic U, Darden D, Fenner B, Wacker J, Efron PA, Liesenfeld O, Sweeney TE, Moldawer LL (2021) A Transcriptomic Severity Metric That Predicts Clinical Outcomes in Critically Ill Surgical Sepsis Patients. Crit Care Explor 3:e0554. https://doi.org/10.1097/CCE.0000000000000554
Safarika A, Wacker JW, Katsaros K, Solomonidi N, Giannikopoulos G, Kotsaki A, Koutelidakis IM, Coyle SM, Cheng HK, Liesenfeld O et al (2021) A 29-mRNA host response test from blood accurately distinguishes bacterial and viral infections among emergency department patients. Intensive Care Med Exp 9:31. https://doi.org/10.1186/s40635-021-00394-8
Heidelberg University Hospital, Heidelberger Manual der Lebertransplantation. 2006
Decker SO, Hildebrand D, Bruckner T, Lichtenstern C, Heeg K, Weigand MA, Brenner T, Uhle F (2020) Delta-like Canonical Notch Ligand 1 in Patients Following Liver Transplantation-A Secondary Analysis of a Prospective Cohort Study. Diagn (Basel) 10:11. https://doi.org/10.3390/diagnostics10110894
Decker SO, Kruger A, Wilk H, Grumaz S, Vainshtein Y, Schmitt FCF, Uhle F, Bruckner T, Zimmermann S, Mehrabi A et al (2019) New approaches for the detection of invasive fungal diseases in patients following liver transplantation-results of an observational clinical pilot study. Langenbecks Arch Surg 404:309–325. https://doi.org/10.1007/s00423-019-01769-y
Bassetti M, Azoulay E, Kullberg BJ, Ruhnke M, Shoham S, Vazquez J, Giacobbe DR, Calandra T (2021) EORTC/MSGERC Definitions of Invasive Fungal Diseases: Summary of Activities of the Intensive Care Unit Working Group. Clin Infect Dis 72:S121–S127. https://doi.org/10.1093/cid/ciaa1751
Hospital toolkit for adult sepsis surveillance. Centers for Disease Control and Prevention website. https://www.cdc.gov/sepsis/pdfs/Sepsis-Surveillance-Toolkit-Mar-2018_508.pdf. Published March 2018. Accessed March 1, 2024
He YD, Wohlford EM, Uhle F, Buturovic L, Liesenfeld O, Sweeney TE (2021) The optimization and biological significance of a 29-Host-Immune-mRNA panel for the diagnosis of acute infections and sepsis. J Pers Med 11:735. https://doi.org/10.3390/jpm11080735
Samsudin I, Vasikaran SD (2017) Clinical Utility and Measurement of Procalcitonin. Clin Biochem Rev 38:59–68
Huang DT, Yealy DM, Filbin MR, Brown AM, Chang CH, Doi Y, Donnino MW, Fine J, Fine MJ, Fischer MA et al (2018) Procalcitonin-Guided Use of Antibiotics for Lower Respiratory Tract Infection. N Engl J Med 379:236–249. https://doi.org/10.1056/NEJMoa1802670
Chan YL, Liao HC, Tsay PK, Chang SS, Chen JC, Liaw SJ (2002) C-reactive protein as an indicator of bacterial infection of adult patients in the emergency department. Chang Gung Med J 25:437–445
Ip M, Rainer TH, Lee N, Chan C, Chau SS, Leung W, Leung MF, Tam TK, Antonio GE, Lui G et al (2007) Value of serum procalcitonin, neopterin, and C-reactive protein in differentiating bacterial from viral etiologies in patients presenting with lower respiratory tract infections. Diagn Microbiol Infect Dis 59:131–136. https://doi.org/10.1016/j.diagmicrobio.2007.04.019
Rainer TH, Chan CP, Leung MF, Leung W, Ip M, Lee N, Cautherley GW, Graham CA, Fuchs D, Renneberg R (2009) Diagnostic utility of CRP to neopterin ratio in patients with acute respiratory tract infections. J Infect 58:123–130. https://doi.org/10.1016/j.jinf.2008.11.007
Dellinger RP, Carlet JM, Masur H, Gerlach H, Calandra T, Cohen J, Gea-Banacloche J, Keh D, Marshall JC, Parker MM et al (2004) Surviving Sepsis Campaign guidelines for management of severe sepsis and septic shock. Crit Care Med 32:858–873. https://doi.org/10.1097/01.ccm.0000117317.18092.e4
Vincent JL, Brealey D, Libert N, Abidi NE, O’Dwyer M, Zacharowski K, Mikaszewska-Sokolewicz M, Schrenzel J, Simon F, Wilks M et al (2015) Rapid Diagnosis of Infection in the Critically Ill, a Multicenter Study of Molecular Detection in Bloodstream Infections, Pneumonia, and Sterile Site Infections. Crit Care Med 43:2283–2291. https://doi.org/10.1097/CCM.0000000000001249
Rath PM, Saner F, Paul A, Lehmann N, Steinmann E, Buer J, Steinmann J (2012) Multiplex PCR for rapid and improved diagnosis of bloodstream infections in liver transplant recipients. J Clin Microbiol 50:2069–2071. https://doi.org/10.1128/JCM.00745-12
Kita Y, Iwaki Y, Noguchi K, Tzakis AG, Todo S, Fung JJ, Starzl TE (1996) Daily serum inflammatory cytokine (tumor necrosis factor-alpha, interleukin-6) monitoring in liver transplantation focusing on allograft rejection: a five-case report. Transplant Proc 28:1237–1240
Sweeney TE, Perumal TM, Henao R, Nichols M, Howrylak JA, Choi AM, Bermejo-Martin JF, Almansa R, Tamayo E, Davenport EE et al (2018) A community approach to mortality prediction in sepsis via gene expression analysis. Nat Commun 9:694. https://doi.org/10.1038/s41467-018-03078-2
Sweeney TE, Shidham A, Wong HR, Khatri P (2015) A comprehensive time-course-based multicohort analysis of sepsis and sterile inflammation reveals a robust diagnostic gene set. Sci Transl Med 7:287ra271. https://doi.org/10.1126/scitranslmed.aaa5993
Sweeney TE, Wong HR, Khatri P (2016) Robust classification of bacterial and viral infections via integrated host gene expression diagnostics. Sci Transl Med 8:346ra391. https://doi.org/10.1126/scitranslmed.aaf7165
Kostaki A, Wacker JW, Safarika A, Solomonidi N, Katsaros K, Giannikopoulos G, Koutelidakis IM, Hogan CA, Uhle F, Liesenfeld O et al (2022) A 29-Mrna Host Response Whole-Blood Signature Improves Prediction of 28-Day Mortality and 7-Day Intensive Care Unit Care in Adults Presenting to the Emergency Department with Suspected Acute Infection and/or Sepsis. Shock 58:224–230. https://doi.org/10.1097/SHK.0000000000001970
Decker SO, Kruger A, Wilk H, Uhle F, Bruckner T, Hofer S, Weigand MA, Brenner T, Zivkovic AR (2022) Concurrent Change in Serum Cholinesterase Activity and Midregional-Proadrennomedullin Level Could Predict Patient Outcome following Liver Transplantation. Biomolecules 12:7. https://doi.org/10.3390/biom12070989
Eyraud D, Ben Ayed S, Tanguy ML, Vezinet C, Siksik JM, Bernard M, Fratea S, Movschin M, Vaillant JC, Coriat P, Hannoun L (2008) Procalcitonin in liver transplantation: are high levels due to donors or recipients? Crit Care 12:R85. https://doi.org/10.1186/cc6942
Cousin VL, Lambert K, Trabelsi S, Galetto-Lacour A, Posfay-Barbe KM, Wildhaber BE, McLin VA (2017) Procalcitonin for infections in the first week after pediatric liver transplantation. BMC Infect Dis 17:149. https://doi.org/10.1186/s12879-017-2234-y
Dong R, Wan B, Lin S, Wang M, Huang J, Wu Y, Wu Y, Zhang N, Zhu Y (2019) Procalcitonin and Liver Disease: A Literature Review. J Clin Transl Hepatol 7:51–55. https://doi.org/10.14218/JCTH.2018.00012
Ducharme J, Self WH, Osborn TM, Ledeboer NA, Romanowsky J, Sweeney TE, Liesenfeld O, Rothman RE (2020) A Multi-mRNA Host-Response Molecular Blood Test for the Diagnosis and Prognosis of Acute Infections and Sepsis: Proceedings from a Clinical Advisory Panel. J Pers Med 10:4. https://doi.org/10.3390/jpm10040266
Razonable RR, Emery VC, the Annual Meeting of the IHMF (2004) Management of CMV infection and disease in transplant patients. Herpes 11:77–86
Shbaklo N, Tandoi F, Lupia T, Corcione S, Romagnoli R, De Rosa FG (2022) Bacterial and Viral Infections in Liver Transplantation: New Insights from Clinical and Surgical Perspectives. Biomedicines 10:7. https://doi.org/10.3390/biomedicines10071561
European Association for the Study of the Liver (2016) EASL Clinical Practice Guidelines: Liver transplantation. J Hepatol 64:433–485. https://doi.org/10.1016/j.jhep.2015.10.006
Pastacaldi S, Teixeira R, Montalto P, Rolles K, Burroughs AK (2001) Hepatic artery thrombosis after orthotopic liver transplantation: a review of nonsurgical causes. Liver Transpl 7:75–81. https://doi.org/10.1053/jlts.2001.22040
Sun HY, Cacciarelli TV, Wagener MM, Singh N (2010) Preemptive therapy for cytomegalovirus based on real-time measurement of viral load in liver transplant recipients. Transpl Immunol 23:166–169. https://doi.org/10.1016/j.trim.2010.06.013
Acknowledgements
We would like to thank Ute Krauser and Jan Pfister for their outstanding technical assistance.
Funding
Open Access funding enabled and organized by Projekt DEAL. The study was carried out with financial resources of the Department of Anesthesiology (Heidelberg University Hospital, Germany). Furthermore, this study received a financial grant from Inflammatix for additional biomarker measurements agreed by a contract between Heidelberg University Hospital and Inflammatix. Inflammatix paid the cost of running the host-response classifier.
Author information
Authors and Affiliations
Contributions
SOD conceived the study, participated in its design and coordination, and helped draft the article. Furthermore, he performed data acquisition and prepared the tables and figures. AH performed data acquisition and was involved in critical revision of the article. OL, NW, FU, JS, AM, FCFS and MAW participated in the design of the study and were involved in critical revision of the article. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Informed consent
Informed consent was obtained from all subjects involved in this study.
Consent for publication
Not applicable.
Clinical trial notation
German Clinical Trials Register: DRKS00023236, Registered 07 October 2020, https://drks.de/search/en/trial/DRKS00023236
Institutional review board statement
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of the Medical Faculty of Heidelberg (TrialCode No. S-693/2020, date of approval: 30 September 2020).
Competing interests
OL and NW are employees and stock options holders of Inflammatix, Inc. FU was previously an employee of Inflammatix. SOD received scientific grants from Heidelberg Foundation of Surgery and B.Braun Foundation, Melsungen, Germany, as well as research support from SphingoTec GmbH and Inflammatic Inc. MAW received grants from Köhler Chemie, DFG and BMBF, consulting fees from B. Braun, Gilead, Mundipharma and Boehringer Ingelheim, and payment or honoraria for lectures, presentations, or educational events from MSD, Gilead, Shionogi, Pfizer and Beckman Coulter. He is also a patent owner (EP17185036.5 and EP17198330.7), has participated on advisory boards for MSD, Gilead, Shionogi, Biotest, Pfizer, Eumedica, SOBI and Beckman Coulter, is the vice-head of the German Sepsis Society and member of the scientific advisory council of PEG, and is cofounder of Delta Theragnostics. The following authors declare no conflicts of interest: JS, FCFS, AH, AM.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Halder, A., Liesenfeld, O., Whitfield, N. et al. A 29-mRNA host-response classifier identifies bacterial infections following liver transplantation – a pilot study. Langenbecks Arch Surg 409, 185 (2024). https://doi.org/10.1007/s00423-024-03373-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00423-024-03373-1