Abstract
Purpose
Iron is an important component of the oxygen-binding proteins and may be critical to optimal athletic performance. Previous studies have suggested that the G allele of C/G rare variant (rs1799945), which causes H63D amino acid replacement, in the HFE is associated with elevated iron indexes and may give some advantage in endurance-oriented sports. The aim of the present study was to investigate the association between the HFE H63D polymorphism and elite endurance athlete status in Japanese and Russian populations, aerobic capacity and to perform a meta-analysis using current findings and three previous studies.
Methods
The study involved 315 international-level endurance athletes (255 Russian and 60 Japanese) and 809 healthy controls (405 Russian and 404 Japanese). Genotyping was performed using micro-array analysis or by PCR. VO2max in 46 male Russian endurance athletes was determined using gas analysis system.
Results
The frequency of the iron-increasing CG/GG genotypes was significantly higher in Russian (38.0 vs 24.9%; OR 1.85, P = 0.0003) and Japanese (13.3 vs 5.0%; OR 2.95, P = 0.011) endurance athletes compared to ethnically matched controls. The meta-analysis using five cohorts (two French, Japanese, Spanish, and Russian; 586 athletes and 1416 controls) showed significant prevalence of the CG/GG genotypes in endurance athletes compared to controls (OR 1.96, 95% CI 1.58–2.45; P = 1.7 × 10–9). Furthermore, the HFE G allele was associated with high V̇O2max in male athletes [CC: 61.8 (6.1), CG/GG: 66.3 (7.8) ml/min/kg; P = 0.036].
Conclusions
We have shown that the HFE H63D polymorphism is strongly associated with elite endurance athlete status, regardless ethnicities and aerobic capacity in Russian athletes.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Iron is an important component of the oxygen-binding proteins, such as hemoglobin and myoglobin. Whereas hemoglobin transports oxygen (via erythrocytes), myoglobin’s function is to store oxygen in working skeletal muscles and to facilitate its transport to mitochondria. Approximately 65% of iron is stored in hemoglobin (Wallace 2016), thus there is a positive correlation between serum iron concentrations and hemoglobin (Ofojekwu et al. 2013; Baart et al. 2018). Iron can affect many physiological processes, and its deficiency is associated with fatigue, anemia, and decreased exercise performance (DellaValle 2013; Abbaspour et al. 2014). There is a balance between iron loss, iron absorption, and iron storage to maintain iron homeostasis (DellaValle 2013; Wallace 2016; Rubeor et al. 2018). Endurance athletes have an increased risk for iron loss because of the insufficient dietary intake and training intensity, which leads to increased risk for suboptimal iron status (Hinton 2014).
Serum iron measures and hematological parameters have significant heritability components. The heritability estimates are 23% for iron, 29–37% for ferritin, and 28% for transferrin saturation (Njajou et al. 2006; McLaren et al. 2010), and 84% for hemoglobin (Evans et al. 1999). Genetic variation plays a significant role in interindividual differences in serum iron parameters. More specifically, previous studies have suggested that the missense mutations of the hemochromatosis (HFE) gene are associated with iron indexes (Burt et al. 1998; Wallace 2016). The proportion of variance explained by HFE gene mutations was reported to be 2.1% for serum iron level, 5.6% for ferritin, and 3.5% for transferrin saturation (Njajou et al. 2006).
The HFE gene (full name—homeostatic iron regulator) is a protein coding gene located on chromosome 6. The protein regulates iron absorption by regulating the interaction of the transferrin receptor with transferrin. The HFE protein interacts with TFRC, the transferrin receptor, so its primary mode of action is through regulation of the iron storage hormone hepcidin. Individuals with one (C/G or H63D genotype) or two (G/G or D63D genotype) missense mutations of the H63D (also known as His63Asp or rs1799945 C/G) polymorphism, show higher circulating iron concentrations than people without mutations (Burt et al. 1998). In the H63D carrier group, a positive correlation between iron and hemoglobin was noted (Barbara et al. 2016). The H63D mutation is commonly found in European (17%) and American (12%) populations, and is rarer in East Asian (3%), South Asian (7%), and African (1%) populations.
The H63D mutation accounts for a mild form of hereditary hemochromatosis (HH), a condition with increased intestinal iron absorption which may lead to liver fibrosis and cirrhosis, hepatocellular carcinoma, diabetes mellitus, cardiomyopathy, and hypogonadotropic hypogonadism (Wallace 2016). Given the importance of iron and hemoglobin in athletic performance, one might suggest that the HFE gene H63D may give some advantage in endurance sports. Indeed, Deugnier et al. (2002) have identified an increased frequency of the G allele in 77 elite French road male cyclists when compared to controls (24.7 vs 17.1%, P = 0.04). In accordance with this data, the frequency of the C/G genotype was significantly higher in 65 professional Spanish endurance athletes (50 road cyclists and 15 endurance runners) in comparison with controls (41.5 vs 24.6%, P = 0.01) (Chicharro et al. 2004). Finally, Hermine et al. (2015) have found that the frequency of CG/GG genotypes was significantly higher in the group of French elite athletes compared to controls (38% vs 21.9%, P = 0.0019). Although these findings in West European populations support the hypothesis that the iron-increasing HFE rs1799945 G allele is favorable for endurance performance, replication studies in different ethnic groups using different designs are warranted. This approach leads to the exclusion of false-positive genetic associations (Eynon et al. 2013; Zarebska et al. 2017; Papadimitriou et al. 2018; Yvert et al. 2018; Guilherme et al. 2019; Pickering et al. 2019).
The aim of the study was to investigate the association between the HFE gene H63D polymorphism and endurance athlete status in Japanese and Russian populations, aerobic capacity, and to perform a meta-analysis using current findings and three previous studies.
Methods
Ethical approval
The study was approved by the Ethics Committee of the Physiological Section of the Russian National Committee for Biological Ethics, Ethics Committees of the Juntendo University and National Institutes of Biomedical Innovation, Health and Nutrition (Japan) and by the Institutional Research Board of Anti-Doping Laboratory Qatar (ADLQ) (F2014000009). Written informed consent was obtained from each participant. The study complied with the guidelines set out in the World Medical Association Declaration of Helsinki and ethical standards in sport and exercise science research. The experimental procedures were conducted in accordance with the set of guiding principles for reporting the results of genetic association studies defined by the Strengthening the reporting of genetic association studies (STREGA) statement.
Study participants
The study involved 315 international-level endurance athletes (255 Russian and 60 Japanese) and 809 healthy controls (405 Russian and 404 Japanese). The first group comprised 255 international-level Russian endurance athletes tested negative for doping substances and involved in biathlon, kayaking, cross-country skiing, cycling, rowing, running ≥ 800 m, speed skating ≥ 1.5 km, swimming ≥ 400 m, and triathlon. Controls were 405 healthy, unrelated citizens of Russia without any competitive sport experience. Of those, 46 male endurance athletes (middle-distance athletes (n = 31): rowers, kayakers, speed skaters; long-distance athletes (n = 15): biathletes and cross-country skiers) participated in the study of aerobic performance. The second group involved 60 Japanese international-level endurance athletes (800 m to marathon runners) tested negative for doping substances, including several world record holders and medallist in Olympic Games. Controls were (n = 406) healthy, unrelated citizens of Japanese.
Genotyping
DNA samples of the Russian cohorts were majorly genotyped using micro-array analysis, as described previously (Pickering et al. 2019). In part, some DNA samples of Russian athletes and controls were genotyped for the HFE rs1799945 polymorphism with a TaqMan® SNP Genotyping Assay (Thermo Fisher Scientific Inc, Waltham, Massachusetts, USA) with a StepOne TM Real-Time PCR System (Thermo Fisher Scientific Inc, Waltham, Massachusetts, USA) or using PCR-restriction fragment length polymorphism (RFLP) method, according to a previously described method (Merryweather-Clarke et al. 1997).
Japanese cohort: total DNA was extracted from saliva or venous blood using Oragene DNA Collection Kit (DNA genotek, Ontario, Canada) or QIAamp DNA blood Maxi Kit (QIAGEN, Hilden, Germany), respectively. Illumina® HumanOmniExpress Beadchip (Illumina Inc, Hayward, California, USA) were used for genotyping of more than 700,000 SNPs in athletes and controls. The genotype calls were performed with Illumina® GenomeStudio Software. Genotype data of the HFE rs1799945 polymorphism were obtained from the genotyping results of the Illumina® HumanOmniExpress Beadchip.
VO2max measurement
Maximal oxygen consumption rate (V̇O2max) in rowers was determined using an incremental test to exhaustion on a PM 3 rowing ergometer (Concept II, Morrisville, Vermont, USA). The initial workload was 150 W. The duration of exercise at each workload was 3 min, with a 30 s rest period between increments of 50 W. VO2 and VCO2 was determined breath by breath by a MetaMax 3B gas analysis system (Cortex, Leipzig, Germany) using an electro-chemical cell and non-dispersive infrared sensor, respectively; air flow was measured using a turbine transducer (Triple V). Two-point gas calibrations (first gas—15% O2, 5% CO2; second gas—ambient air) were performed daily. A one-point gas calibration with ambient air was performed before each test as well as a flow transducer calibration using a 3 L syringe (Hans Rudolph, Kansas City, USA). The criteria used to confirm a maximal test were a decrease in power of more than 30 W from the target power despite strong verbal encouragement and a respiratory exchange ratio greater than 1.1 before cessation of exercise. V̇O2max was recorded as the highest mean value observed over a 30 s period.
V̇O2max in kayakers was determined using an incremental test to exhaustion on a kayaking ergometer (Efremov, Moscow, Russia). The initial workload was 8 kg for men and 5 kg for women. The duration of exercise at each workload was 2 min, with a 30 s rest period between increments of 1 kg. V̇O2max was determined breath by breath using a MetaLyzer II gas analysis system (Cortex Biophysik, Leipzig, Germany). VO2max was recorded as the highest mean value observed over a 30 s period.
V̇O2max in speed skaters was determined using a ramp test to exhaustion on an electromagnetic cycle ergometer Ergoselect 200 K (Ergoline, Bitz, Germany). The initial workload was 60 W, the increment was 15 W/min, and the target cadence was 60–70 rpm. V̇O2max was determined breath by breath using a MetaMax 3B gas analysis system (Cortex Biophysik, Leipzig, Germany). The criteria used to confirm a maximal test were a decrease in cadence of less than 50 rpm despite strong verbal encouragement and a respiratory exchange ratio greater than 1.1 before cessation of exercise. V̇O2max was recorded as the highest mean value observed over a 30 s period.
V̇O2max in biathletes and cross-country skiers was determined using an incremental test to exhaustion on a treadmill HP Cosmos (h/p/cosmos sports & medical gmbh, Nussdorf, Germany). The initial speed was 7 km/h, the increment was 0.1 km/h every 10 s. V̇O2max was determined breath by breath using a MetaMax 3B-R2 gas analysis system (Cortex Biophysik, Leipzig, Germany). V̇O2max was recorded as the highest mean value observed over a 30 s period.
Selection of studies for the meta-analysis
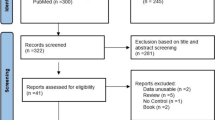
Databases of PubMed, Web of Science, Science Direct and Google Scholar were searched for association studies as of July 19, 2019. The terms used were “HFE” and “athletes” restricted to English. The exclusion criteria were: (1) review; (2) non-English; (3) studies did not involve endurance athletes; (4) ethnically mixed group of athletes were analysed (given that allelic frequencies vary significantly across different ethnicities; for example, we did not include the study of Grealy et al. (2015) because the mixed group of athletes from North America, Europe, Oceania, South America, Asia, and Africa was studied); and (5) duplicates. The inclusion criteria were: (1) case–control study design evaluating the association between HFE gene H63D polymorphism and endurance athlete status; (2) sufficient genotype frequency data to calculate the odds ratios (ORs) and 95% confidence intervals (CIs) and (3) athletes and controls in studies comply with the Hardy–Weinberg equilibrium (HWE). Overall, seven articles published between 1998 and 2015 were identified of which three were found as eligible including a total number of 271 endurance athletes and 607 controls.
Statistical analysis
Genotype distribution and allele frequencies between athletes and controls were compared using χ2 tests. Differences in phenotype between groups were analysed using unpaired t tests. Data are presented as mean (standard deviation). Statistical analyses were conducted using GraphPad InStat software (GraphPad Software, Inc., California, USA) and PLINK software program (Purcell et al. 2007). To perform the meta-analysis with obtained data and all published studies the Cochrane Review Manager (RevMan) (London, UK) version 5.3 was used. Random and fixed effect models were applied. Odds ratio with 95% confidence intervals (CI) was estimated using the Mantel–Haenszel method. The heterogeneity degree between the studies was assessed with the I2 statistics. P values < 0.05 were considered statistically significant.
Results
Case–control study
In Japanese and Russian groups of athletes and controls, the HFE gene rs1799945 polymorphism met Hardy–Weinberg expectations (P > 0.05 in both groups tested separately). The frequencies of the rs1799945 G allele were significantly higher in Russian (21.0 vs 13.2%; P = 0.0002) and Japanese (7.5 vs 2.5%; P = 0.0032) endurance athletes compared to ethnically matched controls (Table 1). Furthermore, the rs1799945 CG/GG genotypes were significantly over- represented in Russian (38.0 vs 24.9%; OR 1.85, P = 0.0003) and Japanese (13.3 vs 5.0%; OR 2.95, P = 0.011) endurance athletes compared to ethnically matched controls (Table 2). These results remained statistically significant after correction for multiple testing.
Meta-analysis
Multi-database literature search yielded three eligible studies involving endurance athletes that were genotyped for the HFE gene H63D polymorphism. These involved 77 French elite road cyclists and 254 controls (Deugnier et al. 2002); 65 Spanish highly trained athletes (50 professional road cyclists and 15 Olympic class endurance runners) and 134 controls (sedentary men from Spain) (Chicharro et al. 2004) and 129 French elite athletes (Nordic skiing, rowing, fighting) and 219 controls (Hermine et. al. 2015). The genotypic frequencies for both the cases and the controls in all studies were in Hardy–Weinberg equilibrium.
The frequencies of the rs1799945 CG/GG genotypes were significantly higher in three groups of French and Spanish endurance athletes compared to controls (Table 2). Overall, five case–control studies (two current and three previous) including a total number of 586 endurance athletes and 1416 controls were used for the meta-analysis. The pooled OR for the CG/GG genotypes compared to the CC genotype was 1.95 (95% CI 1.57–2.43, P = 2.5 × 10–9 for the fixed effect model) and 1.96 (95% CI 1.58–2.45, P = 1.7 × 10–9 for the random effect model) (Fig. 1). No heterogeneity between studies (I2 = 0%; P = 0.83) was observed. These results indicate that the carriage of the HFE mutation (i.e. CG/GG genotypes) is strongly associated with endurance athlete status.
Aerobic study
We identified that the HFE gene rs1799945 G allele was significantly associated with increased V̇O2max in the whole group of Russian male endurance athletes (CC [n = 29]: 61.8 (6.1), CG/GG [n = 17]: 66.3 (7.8) ml/min/kg; P = 0.036), as well as in long-distance athletes only (CC [n = 8]: 68.1 (3.4), CG/GG [n = 7]: 73.0 (4.6) ml/min/kg; P = 0.038).
Discussion
This is the first study to demonstrate that H63D variation in the HFE is associated with elite endurance athlete status in Russian and Japanese populations. More specifically, we found that the frequencies of the iron-increasing genotypes (i.e. CG/GG) were significantly higher in Russian and Japanese elite endurance athletes compared to ethnically matched controls. We also confirmed the observation that the H63D mutation is commonly found in East Europeans (13.2%) and is rarer in East Asian (2.5%) populations. In addition, the meta-analysis using five cohorts (two French, Japanese, Spanish, and Russian) including a total number of 586 endurance athletes and 1416 controls showed significantly higher prevalence of the CG/GG genotypes in endurance athletes compared with controls.
The H63D polymorphism is functional given that the rare G allele has been shown to reduce the ability of the HFE protein to bind to its ligand, thereby preventing the inhibition of transferrin–TFRC binding and resulting in increased transport of iron into circulation and cells (Feder et al. 1998). The hypothesis that the iron-increasing HFE rs1799945 G (63D) allele is favorable for endurance performance was confirmed in our functional study, where we identified that the G allele was associated with increased V̇O2max in Russian male endurance athletes. One might suggest that the favorable effect of the HFE G allele on aerobic capacity and ability to become an endurance athlete is mediated through its impact on hematological parameters, as was shown in the study of French endurance athletes (Hermine et al. 2015). Furthermore, in the genome-wide association study (GWAS) of 173,480 European-ancestry participants, the HFE rs1799945 G allele was shown to be significantly (P < 5 × 10–8) associated with increased values of hematological parameters, such as hematocrit, mean corpuscular hemoglobin concentration, hemoglobin, and reticulocyte count (Astle et al. 2016). Previous studies in athletes have also shown that variations in genes, which regulate hematological traits, are associated with aerobic capacity and endurance athlete status (Ahmetov et al. 2015; Malczewska-Lenczowska et al. 2016).
Our findings seem reasonable given the importance of iron metabolism for endurance athletes (Abbaspour et al. 2014). The leading role of iron is to transport oxygen into the red blood cells and tissues, and it does so mainly through hemoglobin. Furthermore, iron is present in myoglobin and cytochromes of skeletal muscle mostly in oxidative (slow-twitch) muscle fibers. The normal level of iron is crucial to maintain redox balance in muscle and produce mitochondrial energy production, which are primary factors determining exercise performance (Buratti et al. 2015). Iron deficiency without anemia and/or sports anemia are a common issue in athletic populations (at 15–35% of female and 3–11% of male athletes) (Fallon 2008; Malczewska et al. 2001; Parks et al. 2017) with higher frequency in endurance athletes, e.g., distance runners and triathletes (Rietjens et al. 2002; Lukaski 2004; Sinclair and Hinton 2005), and physically active individuals compared with sedentary controls (Milic et al. 2011; Eliakim et al. 2002; Gropper et al. 2006; Woolf et al. 2009). Factors, which could affect both male and female athletes’ iron stores, is low energy intake, inadequate dietary iron intake, vegetarian diets, exercise-associated iron losses, reduced iron recycling (Hinton 2014; Castell et al. 2019; Sim et al. 2019), and menstrual blood losses in female athletes (Pedlar et al. 2018). Iron deficiency, which is accompanied by a reduction of oxygen transport to the working skeletal muscle, can lead to lower blood pH, depletion of muscle glycogen, which may negatively affect the endurance performance and exercise economy (Sim et al. 2019).
Humans with experimentally induced anemia showed reduced VO2max, which is proportional to hemoglobin concentrations (Woodson et al. 1978; Celsing et al. 1986). Iron supplementation of anemic women improved iron status and performance during a standardized, multi-stage treadmill test and reduced exercise heart rate and blood lactate concentrations (Gardner et al. 1975). Although iron supplementation does not necessarily improve VO2max (Klingshirn et al. 1992; Zhu and Haas 1998), this strategy is useful for iron-deficient nonanemic athletes in the improvement of athletic performance in endurance sports (Burden et al. 2015; Rubeor et al. 2018). Therefore, the iron status of athletes should be monitored systematically throughout the training and competition season to early detection or prevention of iron deficiency.
The limitation of our study is the small sample sizes of Japanese athletes, as well as sub-group of Russian athletes with VO2max data, which may lead to potential type I errors. As in all such studies, extension to, and replication within other racial groups is proposed.
In conclusion, we have shown that the HFE gene H63D polymorphism is strongly associated with endurance athlete status across East Asian, East and West European populations and with aerobic capacity in Russian athletes.
Abbreviations
- CI:
-
Confidence intervals
- DNA:
-
Deoxyribonucleic acid
- EDTA:
-
Ethylenediaminetetraacetic acid
- GWAS:
-
Genome-wide association studies
- HFE:
-
Homeostatic iron regulator (hemochromatosis gene)
- HH:
-
Hereditary hemochromatosis
- HWE:
-
Hardy–Weinberg equilibrium
- PCR:
-
Polymerase chain reaction
- RFLP:
-
Restriction fragment length polymorphism
- SNP:
-
Single-nucleotide polymorphism
- STREGA:
-
Strengthening the reporting of genetic association
- TFRC:
-
Transferrin receptor
- V̇O2max :
-
Maximal oxygen consumption
References
Abbaspour N, Hurrell R, Kelishadi R (2014) Review on iron and its importance for human health. J Res Med Sci 19(2):164–174
Ahmetov I, Kulemin N, Popov D, Naumov V, Akimov E, Bravy Y, Egorova E, Galeeva A, Generozov E, Kostryukova E, Larin A, Lj M, Ospanova E, Pavlenko A, Starnes L, Żmijewski P, Alexeev D, Vinogradova O, Govorun V (2015) Genome-wide association study identifies three novel genetic markers associated with elite endurance performance. Biol Sport 32(1):3–9
Astle WJ, Elding H, Jiang T, Allen D, Ruklisa D, Mann AL, Mead D, Bouman H, Riveros-Mckay F, Kostadima MA, Lambourne JJ, Sivapalaratnam S, Downes K, Kundu K, Bomba L, Berentsen K, Bradley JR, Daugherty LC, Delaneau O, Freson K, Garner SF, Grassi L, Guerrero J, Haimel M, Janssen-Megens EM, Kaan A, Kamat M, Kim B, Mandoli A, Marchini J, Martens JHA, Meacham S, Megy K, O'Connell J, Petersen R, Sharifi N, Sheard SM, Staley JR, Tuna S, van der Ent M, Walter K, Wang SY, Wheeler E, Wilder SP, Iotchkova V, Moore C, Sambrook J, Stunnenberg HG, Di Angelantonio E, Kaptoge S, Kuijpers TW, Carrillo-de-Santa-Pau E, Juan D, Rico D, Valencia A, Chen L, Ge B, Vasquez L, Kwan T, Garrido-Martín D, Watt S, Yang Y, Guigo R, Beck S, Paul DS, Pastinen T, Bujold D, Bourque G, Frontini M, Danesh J, Roberts DJ, Ouwehand WH, Butterworth AS, Soranzo N (2016) The allelic landscape of human blood cell trait variation and links to common complex disease. Cell 167(5):1415–1429
Baart AM, Balvers MGJ, Hopman MTE, Eijsvogels TMH, Klein Gunnewiek JMT, van Kampen CA (2018) Reticulocyte hemoglobin content in a large sample of the general Dutch population and its relation to conventional iron status parameters. Clin Chim Acta 483:20–24
Barbara KH, Marcin L, Jedrzej A, Wieslaw Z, Elzbieta AD, Malgorzata M, Ewa M, Jacek KJ (2016) The impact of H63D HFE gene carriage on hemoglobin and iron status in children. Ann Hematol 95(12):2043–2048
Buratti P, Gammella E, Rybinska I, Cairo G, Recalcati S (2015) Recent advances in iron metabolism: relevance for health, exercise, and performance. Med Sci Sports Exerc 47(8):1596–1604
Burden RJ, Morton K, Richards T, Whyte GP, Pedlar CR (2015) Is iron treatment beneficial in, iron-deficient but non-anaemic (IDNA) endurance athletes? A systematic review and meta-analysis. Br J Sports Med 49(21):1389–1397
Burt MJ, George PM, Upton JD, Collett JA, Frampton CM, Chapman TM, Walmsley TA, Chapman BA (1998) The significance of haemochromatosis gene mutations in the general population: implications for screening. Gut 43:830–836
Castell LM, Nieman DC, Bermon S, Peeling P (2019) Exercise-induced illness and inflammation: can immunonutrition and iron help? Int J Sport Nutr Exer Metab 29(2):181–188
Celsing F, Blomstrand E, Werner B, Pihlstedt P, Ekblom B (1986) Effects of iron deficiency on endurance and muscle enzyme activity in man. Med Sci Sports Exerc 18(2):156–161
Chicharro JL, Hoyos J, Gómez-Gallego F, Villa JG, Bandrés F, Celaya P, Jiménez F, Alonso JM, Córdova A, Lucia A (2004) Mutations in the hereditary haemochromatosis gene HFE in professional endurance athletes. Br J Sports Med 38:418–421
DellaValle DM (2013) Iron supplementation for female athletes: effects on iron status and performance outcomes. Curr Sports Med Rep 12(4):234–239
Deugnier Y, Loréal O, Carré F, Duvallet A, Zoulim F, Vinel JP, Paris JC, Blaison D, Moirand R, Turlin B, Gandon Y, David V, Mégret A, Guinot M (2002) Increased body iron stores in elite road cyclists. Med Sci Sports Exerc 34:876–880
Eliakim A, Nemet D, Constantini N (2002) Screening blood tests in members of the Israeli National Olympic team. J Sports Med Phys Fit 42(2):250–255
Evans DM, Frazer IH, Martin NG (1999) Genetic and environmental causes of variation in basal levels of blood cells. Twin Res 2:250–257
Eynon N, Nasibulina ES, Banting LK, Cieszczyk P, Maciejewska-Karlowska A, Sawczuk M, Bondareva EA, Shagimardanova RR, Raz M, Sharon Y, Williams AG, Ahmetov II, Lucia A, Birk R (2013) The FTO A/T polymorphism and elite athletic performance: a study involving three groups of European athletes. PLoS ONE 8(4):e60570
Fallon KE (2008) Screening for haematological and iron-related abnormalities in elite athletes-analysis of 576 cases. J Sci Med Sport 11(3):329–336
Feder JN, Penny DM, Irrinki A, Lee VK, Lebrón JA, Watson N, Tsuchihashi Z, Sigal E, Bjorkman PJ, Schatzman RC (1998) The hemochromatosis gene product complexes with the transferrin receptor and lowers its affinity for ligand binding. Proc Natl Acad Sci USA 95(4):1472–1477
Gardner GW, Edgerton VR, Barnard RJ, Bernauer EM (1975) Cardiorespiratory hematological and physical performance responses of anemic subjects to iron treatment. Am J Clin Nutr 28(9):982–988
Grealy R, Herruer J, Smith CL, Hiller D, Haseler LJ, Griffiths LR (2015) Evaluation of a 7-gene genetic profile for athletic endurance phenotype in ironman championship triathletes. PLoS ONE 10(12):e0145171
Gropper SS, Blessing D, Dunham K, Barksdale JM (2006) Iron status of female collegiate athletes involved in different sports. Biol Trace Elem Res 109(1):1–14
Guilherme JPLF, Egorova ES, Semenova EA, Kostryukova ES, Kulemin NA, Borisov OV, Khabibova SA, Larin AK, Ospanova EA, Pavlenko AV, Lyubaeva EV, Popov DV, Lysenko EA, Vepkhvadze TF, Lednev EM, Govorun VM, Generozov EV, Ahmetov II, Lancha Junior AH (2019) The A-allele of the FTO gene rs9939609 polymorphism is associated with decreased proportion of slow oxidative muscle fibers and over-represented in heavier athletes. J Strength Cond Res 33(3):691–700
Hermine O, Dine G, Genty V, Marquet LA, Fumagalli G, Tafflet M, Guillem F, Van Lierde F, Rousseaux-Blanchi MP, Palierne C, Lapostolle JC, Cervetti JP, Frey A, Jouven X, Noirez P, Toussaint JF (2015) Eighty percent of French sport winners in Olympic, World and Europeans competitions have mutations in the hemochromatosis HFE gene. Biochimie 119:1–5
Hinton PS (2014) Iron and the endurance athlete. Appl Physiol Nutr Metab 39(9):1012–1018
Klingshirn LA, Pate RR, Bourque SP, Davis JM, Sargent RG (1992) Effect of iron supplementation on endurance capacity in iron-depleted female runners. Med Sci Sports Exerc 24(7):819–824
Lukaski HC (2004) Vitamin and mineral status: effects on physical performance. Nutrition 20(7–8):632–644
Malczewska J, Szczepańska B, Stupnicki R, Sendecki W (2001) The assessment of frequency of iron deficiency in athletes from the transferrin receptor-ferritin index. Int J Sports Nutr Exer Metab 11(1):42–52
Malczewska-Lenczowska J, Orysiak J, Majorczyk E, Zdanowicz R, Szczepańska B, Starczewski M, Kaczmarski J, Dybek T, Pokrywka A, Ahmetov II, Sitkowski D (2016) Total hemoglobin mass, aerobic capacity, and HBB gene in Polish road cyclists. J Strength Cond Res 30(12):3512–3519
McLaren CE, Barton JC, Eckfeldt JH, McLaren GD, Acton RT, Adams PC, Henkin LF, Gordeuk VR, Vulpe CD, Harris EL, Harrison BW, Reiss JA, Snively BM (2010) Heritability of serum iron measures in the hemochromatosis and iron overload screening (HEIRS) family study. Am J Hematol 85(2):101–105
Merryweather-Clarke AT, Pointon JJ, Shearman JD, Robson KJ (1997) Global prevalence of putative haemochromatosis mutations. J Med Genet 34(4):275–278
Milic R, Martinovic J, Dopsaj M, Dopsaj V (2011) Haematological and iron-related parameters in male and female athletes according to different metabolic energy demands. Eur J Appl Physiol 111(3):449–458
Njajou OT, Alizadeh BZ, Aulchenko Y, Zillikens MC, Pols HA, Oostra BA, Swinkels DW, van Duijn CM (2006) Heritability of serum iron, ferritin and transferrin saturation in a genetically isolated population, the Erasmus Rucphen Family (ERF) study. Hum Hered 61(4):222–228
Ofojekwu M-JN, Nnanna OU, Okolie CE, Odewumi LA, Isiguzoro IOU, Lugos MD (2013) Hemoglobin and serum iron concentrations in menstruating nulliparous women in Jos, Nigeria. Lab Med 44(2):121–124
Papadimitriou ID, Lockey SJ, Voisin S, Herbert AJ, Garton F, Houweling PJ, Cieszczyk P, Maciejewska-Skrendo A, Sawczuk M, Massidda M, Calò CM, Astratenkova IV, Kouvatsi A, Druzhevskaya AM, Jacques M, Ahmetov II, Stebbings GK, Heffernan S, Day SH, Erskine R, Pedlar C, Kipps C, North KN, Williams AG, Eynon N (2018) No association between ACTN3 R577X and ACE I/D polymorphisms and endurance running times in 698 Caucasian athletes. BMC Genom 19(1):13
Parks RB, Hetzel SJ, Brooks MA (2017) Iron deficiency and anemia among collegiate athletes: a retrospective chart review. Med Sci Sports Exerc 49(8):1711–1715
Pedlar CR, Brugnara C, Bruinvels G, Burden R (2018) Iron balance and iron supplementation for the female athlete: a practical approach. Eur J Sport Sci 18(2):295–305
Pickering C, Suraci B, Semenova EA, Boulygina EA, Kostryukova ES, Kulemin NA, Borisov OV, Khabibova SA, Larin AK, Pavlenko AV, Lyubaeva EV, Popov DV, Lysenko EA, Vepkhvadze TF, Lednev EM, Leońska-Duniec A, Pająk B, Chycki J, Moska W, Lulińska-Kuklik E, Dornowski M, Maszczyk A, Bradley B, Kana-ah A, Cięszczyk P, Generozov EV, Ahmetov II (2019) A genome-wide association study of sprint performance in elite youth football players. J Strength Cond Res 33(9):2344–2351
Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, Maller J, Sklar P, de Bakker PIW, Daly MJ, Sham PC (2007) PLINK: a toolset for whole-genome association and population-based linkage analysis. Am J Hum Genet 81(3):559–575
Rietjens GJ, Kuipers H, Hartgens F, Keizer HA (2002) Red blood cell profile of elite olympic distance triathletes A three-year follow-up. Int J Sports Med 23(6):391–396
Rubeor A, Goojha C, Manning J, White J (2018) Does iron supplementation improve performance in iron-deficient nonanemic athletes? Sports Health 10(5):400–405
Sim M, Garvican-Lewis LA, Cox GR, Govus A, McKay AKA, Stellingwerff T, Peeling P (2019) Iron considerations for the athlete: a narrative review. Eur J Appl Physiol. 119(7):1463–1478
Sinclair LM, Hinton PS (2005) Prevalence of iron deficiency with and without anemia in recreationally active men and women. J Am Diet Assoc 105(6):975–978
Wallace DF (2016) The regulation of iron absorption and homeostasis. Clin Biochem Rev 37(2):51–62
Woodson RD, Wills RE, Lenfant C (1978) Effect of acute and established anemia on O2 transport at rest submaximal and maximal work. J Appl Physiol 44(1):36–43
Woolf K, St Thomas MM, Hahn N, Vaughan LA, Carlson AG, Hinton P (2009) Iron status in highly active and sedentary young women. Int J Sport Nutr Exerc Metab 19(5):519–535
Yvert TP, Zempo H, Gabdrakhmanova LJ, Kikuchi N, Miyamoto-Mikami E, Murakami H, Naito H, Cieszczyk P, Leznicka K, Kostryukova ES, Alexeev DG, Egorova ES, Maciejewska-Skrendo A, Larin AK, Generozov EV, Kulemin NA, Ospanova EA, Pavlenko AV, Sawczuk M, Zmijewski P, Lulinska-Kuklik E, Govorun VM, Miyachi M, Ahmetov II, Fuku N (2018) AGTR2 and sprint/power performance: a case-control replication study for rs11091046 polymorphism in two ethnicities. Biol Sport 35(2):105–109
Zarebska A, Jastrzebski Z, Ahmetov II, Zmijewski P, Cieszczyk P, Leonska-Duniec A, Sawczuk M, Leznicka K, Trybek G, Semenova EA, Maciejewska-Skrendo A (2017) GSTP1 c.313A%3eG polymorphism in Russian and Polish athletes. Physiol Genom 49(3):127–131
Zhu YI, Haas JD (1998) Altered metabolic response of iron-depleted nonanemic women during a 15-km time trial. J Appl Physiol 84(5):1768–1775
Author information
Authors and Affiliations
Contributions
DVP, ESK, VPP, LBA, MAE, EVG, NF, and IIA conceived and designed the study. EAS, EM-M, EBA, FA-K, HM, HZ, NAK, AKL, OVB, MM, DVP, EAB, MK, HK, HN, VPP, DAD, EVL, YEP, NF, and IIA acquired, analyzed, and interpreted data. EAS and IIA drafted the manuscript and all authors reviewed and revised it critically for important intellectual content. All authors approved the final version of the manuscript and agreed to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All persons designated as authors qualify for authorship, and all those who qualify for authorship are listed.
Corresponding author
Ethics declarations
Conflict of interest
The authors report no conflict of interest.
Additional information
Communicated by Michael Lindinger.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Semenova, E.A., Miyamoto-Mikami, E., Akimov, E.B. et al. The association of HFE gene H63D polymorphism with endurance athlete status and aerobic capacity: novel findings and a meta-analysis. Eur J Appl Physiol 120, 665–673 (2020). https://doi.org/10.1007/s00421-020-04306-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00421-020-04306-8