Abstract
Exploring the three-dimensional (3D) morphology of neurons is essential to understanding spinal cord function and associated diseases comprehensively. However, 3D imaging of the neuronal network in the broad region of the spinal cord at cellular resolution remains a challenge in the field of neuroscience. In this study, to obtain high-resolution 3D imaging of a detailed neuronal network in the mass of the spinal cord, the combination of synchrotron radiation micro-computed tomography (SRμCT) and the Golgi-cox staining were used. We optimized the Golgi-Cox method (GCM) and developed a modified GCM (M-GCM), which improved background staining, reduced the number of artefacts, and diminished the impact of incomplete vasculature compared to the current GCM. Moreover, we achieved high-resolution 3D imaging of the detailed neuronal network in the spinal cord through the combination of SRμCT and M-GCM. Our results showed that the M-GCM increased the contrast between the neuronal structure and its surrounding extracellular matrix. Compared to the GCM, the M-GCM also diminished the impact of the artefacts and incomplete vasculature on the 3D image. Additionally, the 3D neuronal architecture was successfully quantified using a combination of SRμCT and M-GCM. The SRμCT was shown to be a valuable non-destructive tool for 3D visualization of the neuronal network in the broad 3D region of the spinal cord. Such a combinatorial method will, therefore, transform the presentation of Golgi staining from 2 to 3D, providing significant improvements in the 3D rendering of the neuronal network.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The spinal cord, which transmits descending and ascending neural signals, is an essential component of the central nervous system. Neurons, as the main functional units of the spinal cord, are responsible for receiving and transmitting signals (Lovinger 2008). Hence, exploring the morphology of neuronal networks is important to understand the normal function of the spinal cord and the pathogenesis of related disorders (Hogstrom et al. 2016; Xu et al. 1993).
Histological staining techniques are widely used in research on neuronal morphology (Tsai et al. 2003). However, these methods mainly rely on two-dimensional (2D) histological sections. Unfortunately, 2D images of neurons can lead to data misinterpretation because they do not capture morphological information in the third dimension (Parekh and Ascoli 2013; Cedola et al. 2017). Although researchers achieved limited 3D imaging of neural structure using confocal microscopy, it is still challenging to visualize the microstructure of large-size samples using confocal microscopy (St Croix et al. 2005). Therefore, neuroscience research needs an imaging tool that can help to reveal the 3D neuronal networks of a broad range of samples.
Synchrotron radiation micro-computed tomography (SRμCT) has high resolution and is particularly suitable for 3D imaging of the microstructure of low-absorbing biomedical samples (Strotton et al. 2018; Saccomano et al. 2018; Tesarova et al. 2019). This technique has been used for 3D neuroimaging of the spinal cord. In our previous studies, SRμCT was proposed for 3D imaging of intramedullary microvessels (Cao et al. 2015, 2017; Hu et al. 2014, 2015; Wu et al. 2018). In addition, Fratini et al. reported that they achieved 3D imaging of the microvascular network and the neurons in the unstained mouse spinal cord using SRμCT (Fratini et al. 2015). Later, Bukreeva. et al. performed a quantitative investigation of the 3D neuronal network in the mouse spinal cord (Bukreeva et al. 2017). However, in these studies, the visualized 3D neurons were not intact, and morphological information of axons and dendrites was almost completely lacking. This is because axons and dendrites have a very similar refractive index to the extracellular matrix, producing insufficient contrast to define the outlines of the neurites. Therefore, it is challenging to distinguish axons and dendrites from the extracellular matrix in the unstained spinal cord tissue. 3D imaging of a single neuron in its entirety is a very desirable research goal. Increasing the contrast of the neuronal features is the key to visualizing the entire 3D architecture of neurons through SRμCT.
The Golgi staining technique, based on the impregnation of neural tissue with a heavy metal precipitate, is a classic neuronal staining technique that provides a high-resolution view, allowing all morphological features of the neuron to be visualized (Kassem et al. 2018; Parekh and Ascoli 2013). The heavy metal precipitate is deposited in the somas, axons, and dendrites of neurons, increasing their absorption contrast from the extracellular matrix, which helps outline individual neurons in their entirety. A previous study reported 3D imaging of the human cortex using a combination of conventional micro-CT and Golgi labeling(Mizutani et al. 2008). However, excessive sample artefacts and incomplete vasculature largely prevented the full 3D structure of single neurons from being clearly identified. Hence, a modified Golgi staining method is necessary for improved 3D visualization of neurons. A commercially available kit that is based on Golgi-Cox impregnation and is widely used in laboratories. However, when the procedure was performed according to the current standard manufacture’s protocol, staining usually results in different degrees of background staining, numerous artefacts, and incomplete vasculature. In the present study, we optimized the Golgi-Cox method (GCM) and developed a modified GCM (M-GCM) that can further clear the background, reduce the density of artefacts, and incomplete vascular structure. Moreover, we further combined the M-GCM and SRμCT to achieve improved 3D imaging of neuronal networks in a mass of the mouse spinal cord. The combinatorial method shown here does neither require a destructive sample preparation procedure, then allowing the 3D imaging of the neuronal network in a mass of mouse spinal cord matrix. It will further promote the progress of the visualization of the neuromorphology.
Material and methods
Experimental animals and ethics statement
All research protocols were approved by the Animal Ethics Committee of Central South University. Animal care and use were conducted under the guidelines of the Administration Committee of Affairs Concerning Experimental Animals in Hunan Province, China. A total of 24 adult male C57/BL6 mice weighing approximately 20–23 g each were obtained from the Animal Center of Central South University and were kept in a temperature-controlled room with a 12/12-h light/dark cycle with food and water ad libitum. All mice were randomly divided into two groups, the GCM group (n = 12) and the M-GCM group (n = 12). In each group, eight mice were used for Golgi staining and SRμCT detection and histology (n = 8). Four mice were used for residual blood comparison (n = 4).
Comparison of fresh spinal cord tissue
To confirm M-GCM could completely remove the blood in vessels, the fresh spinal cord tissue was used for comparison between two groups before the Golgi impregnation. In the GCM group, four experimental mice were deeply anesthetized with ketamine (100 mg/kg, intraperitoneal injection)/ xylazine (10 mg/kg, intraperitoneal injection) and were euthanized using a CO2 chamber (CO2 flow rate: 3 L/min). Continue CO2 until 1 min after breathing stops. After euthanasia, the thoracic spinal cord from four randomly selected mice was directly removed and rinsed in the double-distilled water (dd-H2O). In the M-GCM group, another four randomly selected mice were perfused with 100 ml artificial cerebrospinal fluid after euthanasia (ACSF: dd-H2O 1000 ml, NaCl 125 mM, KCl 3 mM, CaCl2 2.5 mM, MgSO4 1.3 mM, NaH2PO4 1.25 mM, NaHCO3 26 mM, Glucose 13 mM) until blood was utterly flushed out, which took up to 5 min, and then the spinal cord was removed. After that, the fresh spinal cord tissue in the two groups were both sectioned sagittally using a freezing microtome (Leica). The thickness of the sections was 200 μm. Sections were observed using the stereomicroscope and 40 × were captured for comparison.
Golgi-Cox impregnation
The GCM group
An FD Rapid GolgiStain Kit (FD NeuroTechnologies, INC., Catalog: PK401) was used in the present study. Golgi staining was performed according to the current standard protocol (Du 2019). Briefly, the impregnation solution (Solution A/B) was prepared by mixing equal volumes of Solutions A and B at least 24 h before use. Eight experimental mice were euthanized after deeply anesthetized. After euthanasia, we carefully removed the thoracic spinal cord and quickly rinsed the tissue in dd-H2O to remove the blood from the surface. The spinal cord was equally divided into two segments (length: 5 mm; diameter: 1.5 mm). One was used for SRμCT detection and the other for histology. The tissue was immersed in the impregnation solution (Solution A/B, 1:1), and stored at room temperature for 2 weeks in the dark. The impregnation solution was replaced after the first 12 h of immersion or the next day; the tissue was then transferred into Solution C and stored at room temperature in the dark for 72 h. Solution C was replaced at least once after the first 24 h of immersion or the next day. After the above procedures were performed, the spinal cord tissue was dehydrated using a graded ethanol series (70, 80, 90, 95, 100%) at room temperature. Finally, the spinal cord tissue was maintained in 100% ethanol until SRμCT detection. The detailed procedures are shown in the flow chart (Fig. S1).
The M-GCM group
An FD Rapid GolgiStain Kit was used in this group. Eight mice were euthanized using a CO2 chamber as above mentioned. Subsequently, the mice were perfused with 100 ml ACSF. Then the thoracic spinal cord tissue was quickly removed and equally divided into two segments. One was used for SRμCT detection and the other for histology. The tissue was immersed in the impregnation solution (Solution A/B, 1:1) and stored at room temperature for 2 weeks in the dark. The impregnation solution was replaced after the first 12 h. Then, the spinal cord tissue was rinsed in dd-H2O for 24 h. The dd-H2O was replaced after the first 12 h. The spinal cord tissue was transferred into Solution C and stored at room temperature in the dark for 72 h. Solution C was replaced at least once after the first 24 h. Next, the spinal cord tissue was rinsed in dd-H2O for another 24 h. After all the above procedures were completed, the spinal cord tissue was dehydrated using a graded ethanol series (70, 80, 90, 95, 100%) at room temperature. Finally, the spinal cord tissue was maintained in methyl salicylate (Millipore Sigma, M6752-1L) for at least 72 h before SRμCT detection.
Tissue processing and microscopic examination
The Golgi-Cox staining spinal cord tissue was sectioned sagittally or coronally (120 μm) using a freezing microtome (Leica).
The GCM group Three sections from each mouse were selected for histological analysis. The selected sections were mounted on a gelatin-coated microscope slide. Solution D/E was prepared; this solution consisted of one part Solution D, one part Solution E, and two parts dd-H2O. The sections were rinsed twice in dd-H2O two times, placed in the staining solution (Solution D/E) for 5 min, and then rinsed in dd-H2O two more times. The sections were dehydrated in sequential rinses of 50, 75, 95, and 100% ethanol for 4 min each. The sections were then cleared in xylene three times for 5 min each. The sections were coverslipped with Eukitt®quick-hardening mounting medium. An Olympus BX51 microscope equipped with Amscope MU1003 18MP CMOS USB 3.0 digital color camera (Olympus, Tokyo, Japan) was used for microscopic examination. The 40 ×, 100 ×, 200 ×, and 600 × images were captured for analysis. The exposure time was 20–80 ms.
The M-GCM group The selected sections were rinsed in dd-H2O for 5 min, placed in the staining solution (Solution D/E) for 5 min, and then rinsed in dd-H2O for 5 min. The sections were dehydrated in sequential rinses of 50, 75, 95, and 100% ethanol for 4 min each rinse. Next, the sections were mounted on a gelatin-coated microscope slide and left to air-dry for 2 min at room temperature. The sections were cleared in methyl salicylate for half an hour and then coverslipped. The microscopic examination was performed in the same way as for the GCM group.
Comparison of the photomicrograph
Three photomicrographs of sagittal sections were selected from each mouse for comparison (n = 3 × 8).
Stained vasculature comparison: To confirm fewer blood vessels were stained in the M-GCM group, 10 × and 20 × images of sagittal sections (including the area of white matter and gray matter) were captured and compared between the two groups.
Background comparison: To confirm that the M-GCM reduced the background staining, an area with the same size (30 μm × 45 μm) located at the unstained background from the 60 × images in each sample were randomly selected for the comparison of background staining. And the gray value of the background was identified using image J ver. 1.6 (NIH, Bethesda, MD, USA).
Artefact comparison: To confirm the M-GCM generated fewer artefacts (object does not have discernible dendrites or axons), we captured continuous 20 × images (n = 40) located at the gray matter from different sections. The step size was 2 μm. Then the 3D reconstruction was performed using Imaris 9.2 ver. (Imaris Bitplane, Switzerland). Then, artefacts in the 3D volume (740 μm × 900 μm × 80 μm) were manually counted.
X-ray radiation damage analysis
To confirm whether there was X-ray radiation damage on the cellular morphology, we cut the scanned spinal cord into sections. We captured continuous 20 × images and located at the gray matter and performed 3D reconstruction using Imaris 9.2 ver as above mentioned. Then, we performed a comparison of neuronal morphology between scanned tissue and unscanned tissue.
High-resolution SRμCT detection
Specimens were scanned at the BL13W1 beamline of the Shanghai Synchrotron Radiation Facility (SSRF, China). A schematic depiction of beamline experimental station is shown in Fig. 1. The samples were fixed in the middle of the sample stage. The samples were scanned with a photon energy of 20.0 keV. The size of the beam was approximately 45 mm (horizontal) × 5 mm (vertical), and a double-crystal monochromator, with Si (111) and Si (311) crystals, was used to monochromatize the X-rays. After penetration through the sample, X-rays were converted into visible light with a cleaved Lu2SiO5: Ce single-crystal scintillator (10 μm thickness). Projections were magnified by diffraction-limited microscope optics with × 10 magnification and digitized with a high-resolution detector (ORCA Flash 4.0 Scientific CMOS, Hamamatsu K.K., Shizuoka Prefecture, Japan) with a physical pixel size of 0.65 μm × 0.65 μm. The samples were rotated continuously during scanning, and 900 projection images were captured with an angular step size of 0.2° over 180° of rotation. The exposure time was 1 s. The distance between the detector and the sample was adjusted to 3.5 cm. Twenty light-field images and five dark-field images were also collected during each acquisition procedure to correct the electronic noise and variations in the X-ray source brightness.
Schematic depiction of beamline experimental station at the BL13W1 at the Shanghai Synchrotron Radiation Facility (SSRF) in China. The samples were fixed on a sample rotation stage. The images were collected by a detector located at a 3.5 cm distance from the sample after transmission of a monochromatic synchrotron radiation X-ray beam through the sample and delivery to the image acquisition system
Comparison of the projection image
To compare the absorption contrast between the stained neuron and surrounding background from the GCM group and M-GCM group, we selected three corresponding slices from each animal for comparison (n = 3 × 8). A stained neuron at a similar location (near the central canal) from the corresponding slices was selected for comparison. The projection images were processed and analyzed using the Image-Pro Plus (IPP) software program (version 6.0; Media Cybernetics. Bethesda, MD, USA). The gray value (intensity) data were exported from the IPP software.
3D image reconstruction and quantitative determination
All projected tomographic images were transformed into 8-bit slices using the software (Phase-sensitive X-ray Image processing and Tomography Reconstruction, PITRE) developed by the SSRF to perform a direct filtered back-projection algorithm (Chen et al. 2012). Then, all the slices were processed and quantified by Amira software (version 6.01, FEI, USA) (Ian et al. 2016). Depending on the magnitude of X-ray absorption by the neuron, differences in the gray values among tissues were determined. To quantify the neuronal network, we randomly selected the space of 450 × 250 × 1000 μm at the thoracic spinal cord for neuronal network analysis. To compare the neurite length of neurons between the GCM group and M-GCM group, two representative neurons located at the ventral horn (motor neuron) were selected from each mice (n = 2 × 8). And all selected neurons had similar soma volume. Neurite length was identified using Image-Pro Analyzer 3D (version 7.0, Media Cybernetics, Inc., Bethesda, MD, USA) (Yang et al. 2013).
Statistical analysis
All quantitative data are presented as the mean ± standard deviation. All analyses were carried out using SPSS version 24.0 (IBM Corp., Armonk, NY, USA), Groups of data (background gray values, artefact number, gray level deviation of neuronal sites and background, and length of a single neuron neurites) were compared using Student’s t test, and P values less than 0.05 were considered to indicate statistical significance.
Results
M-GCM achieved less background staining, fewer artefacts, and less stained incomplete vasculature compared to the GCM
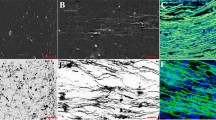
In the GCM group, the residual blood, and Solution A/B left within the spinal cord tissue impacted Golgi staining, leading to high background staining, artefacts, and incomplete vasculature. In the M-GCM group, we found that blood was removed by ACSF perfusion, which led to a few stained incomplete vasculatures, as showed in Fig. 2a–c. In contrast, much blood remained in spinal cord vessels, which results in much stained vasculature in the GCM group (Fig. 2d–f). Besides, the photomicrographs from the M-GCM group and GCM present different degrees of background staining (Fig. 2g, h). The background gray value (213.26 ± 19.58) of the M-GCM group was significantly higher than that (181.52 ± 10.17) of the GCM group (P < 0.0001) (Fig. 2i). Moreover, the 3D images of the spinal cord gray matter from two groups are shown in Fig. 2j, k. The mean number of artefacts in the 3D region from the GCM group and the M-GCM group was 12.12 ± 3.89 and 5.08 ± 2.59, respectively. Artefacts in the M-GCM group were observed to be significantly less (red arrows indicate in Fig. 2k) than the M-GCM group (P < 0.0001) (Fig. 2l). According to the above, slow perfusion with ACSF largely removed blood in the vessels and diminished the interference of vasculature. To sum up, the M-GCM group achieved a more clear background, fewer artefacts, and less incomplete vasculature compared to the GCM group.
Comparison of photomicrographs from the GCM group and M-GCM group. a–c Representative images of the fresh and stained spinal cord tissue M-GCM group (a, b, and c are 4×, 10×, and 20× image, respectively). d–f Representative images of the fresh and stained spinal cord tissue GCM group (d, e, and f are 4×, 10×, and 20× image, respectively). Red arrows indicate the artefacts and the green arrows indicate the incomplete vascular structure. g Representative 60 × image shows low background staining in the M-GCM group. h Representative 60 × image shows high background staining in the GCM group. i A histogram illustrating M-GCM has significantly less the gray value of background compared to that of GCM. j Reconstructive 3D images of neuronal networks in the M-GCM group. k Reconstructive 3D images of neuronal networks in the GCM group. l A histogram illustrating M-GCM has significantly artefact in tissue compared to that of GCM. ***P < 0.0001, Student’s t test was used to determine the statistical significance of the differences [scale bar: 120 μm (g, h) and 250 μm (j, k)]
M-GCM increased the refractive index of the stained spinal cord tissue and achieved high-resolution 3D imaging of neuronal network
In an attempt to achieve 3D imaging of the neuronal network of the spinal cord, the stained spinal cord tissue from the two groups was used for SRμCT scanning. The representative projection images from the M-GCM group and GCM group are shown in Fig. 3a, d, respectively. According to the results, the M-GCM group increased the refractive index of spinal cord tissue compared to the GCM group. The gray level of the line profiles (red line profile) marked in (Fig. 3b, e) was illustrated in (Fig. 3c). In the M-GCM group, the gray level of the unstained area (background) was significantly higher than that of the stained area (the location of the neuron). In contrast, the gray level of the unstained area was similar to that area of the neuronal site in the GCM group. The deviation of the neuronal sites and background in the M-GCM group was 3800.96 ± 565.60, which was significantly more than that (2860.50 ± 472.12) in the GCM group (P < 0.001) (Fig. 3f). Therefore, it was difficult to distinguish the whole neuron and background in the GCM group but M-GCM achieved better. In addition, after these stained spinal cord tissues were detected by SRμCT, numerous 3D neurons could be clearly imaged in the M-GCM group (Fig. 4a). In contrast, there was a large number of artefacts that were difficult to delineate (Fig. 4b) in the GCM group. It was also challenging to outline the entire 3D neuron in the GCM group. 3D images from the GCM group (Fig. 4d) were plagued with more incomplete vasculature compared with 3D images from the M-GCM group (Fig. 4c). According to the above comparison, the M-GCM was further helpful for achieving sufficient contrast to resolve neurons within the tissue and visualizing the 3D neuronal architecture.
Projection images detected by a high-resolution SRμCT. a A representative projection image of a stained spinal cord from the M-GCM group. b Local magnification of the region of interest is denoted by the red frame in a. d A representative projection images of a spinal cord from the GCM group. e Local magnification of the region of interest is denoted by the red frame in d. c Profile of the grey value along the red line in b and e. f A histogram illustrating M-GCM achieved a significantly higher contrast between the neuronal site and surrounding background compared to GCM did. ***P < 0.0001, Student’s t test was used to determine the statistical significance of the differences (scale bar: 200 μm)
M-GCM presented more detailed features of the neuron than the GCM in 3D imaging
The neurons at the ventral horn were selected to show the detailed neuronal network for both groups. As shown in Fig. 5a, the M-GCM group enabled clear visualization of the neuronal network. All entire 3D neurons with axons, dendrites, and soma constituting the neuronal network could be imaged by SRμCT in the M-GCM group (Fig. 5b). But in the GCM group, the incomplete neurons were present in isolation (Fig. 5c). Furthermore, the M-GCM group is better than the GCM group in showing the detailed features of neurons. Based on our schematic illustration of a neuron and photomicrograph demonstrated in Fig. 6a, b, we found that the entire 3D neuronal architecture was visualized in the M-GCM group, as shown in Fig. 6c. However, in the GCM group, only the soma and a small part of neurites were visualized, and most structure of the dendrite and axon was lacking (Fig. 6d). Also, neurons from the M-GCM group had longer neurite length, which ranged from 273.41 μm to 894.36 μm (553.38 ± 177.71 μm), but neurons from the GCM group had short neurite length, which ranged from 78.97 μm to 469.24 μm (210.01 ± 112.06 μm). The average neurite length of a single neuron in the M-GCM group was significantly longer than that in the GCM group (P < 0.001) (Fig. 6e). According to these results, the M-GCM presented more detailed features of the neuron than the GCM did.
Images of the neuronal network in the ventral horn of the mouse spinal cord. a Photomicrograph of the neuronal network in the ventral horn. b Digital 3D image of the neuronal network in the ventral horn from the M-GCM group. c Digital 3D image of the neuronal network in the ventral horn from the GCM group (scale bar: 50 μm)
Images of motor neurons in the mouse spinal cord. a Schematic depiction of a neuron. b Photomicrograph of the motor neurons. c 3D reconstructive motor neurons from the M-GCM group. d 3D reconstructive motor neurons from the GCM group. Red arrows indicate the axons, green arrows indicate dendrites, and yellow arrows indicate soma. A Histogram illustrating M-GCM visualized longed neurite than GCM did. ***P < 0.0001, Student’s t test was used to determine the statistical significance of the differences (scale bar: 50 μm)
Quantification of the 3D neuronal network in M-GCM
After 3D reconstruction, the 3D neuronal network was displayed in Fig. 7a. The highest spatial resolution that we achieved was 2.0 μm. As Fig. 7a shows, somas could be identified and the number of the somas was 76. Regarding the neurons shown in Fig. 7b–d, the length of neurites was 725.32 μm, 489.71 μm, and 267.27 μm, respectively. The result shows that the combination of SRμCT and M-GCM has an advantage in the quantification of detailed neuronal architecture.
a 3D neuronal network at the thoracic mouse spinal cord, quantification for soma number. b–d Neurons with different shapes, measurements of neurites length. As the figure is shown, the combination of SRμCT and M-GCM has an advantage in the quantification of detailed neuronal architecture (scale bar: 50 μm)
Analysis of X-ray damage on cellular morphology
The representative images of neuronal morphology from unscanned spinal cord tissue and scanned spinal cord tissue were displayed in Fig. 8a, b, respectively. There was no significant difference in neuronal morphology between unscanned tissue and scanned tissue. No obvious X-ray radiation damage on cellular morphology was observed.
Discussion
In the present study, M-GCM achieved clearer imaging of neuronal morphology in the spinal cord than GCM did. Furthermore, we achieved high-resolution 3D visualization of entire neurons and quantification of the 3D neuronal network in the mouse spinal cord through a combination of SRμCT and the M-GCM. SRμCT show to be a powerful tool that can visualize and evaluate the 3D morphology of Golgi-Cox stained neurons. In contrast to optical 3D techniques, the approach shown here does not require tissue slicing and allows the investigation of numerous neurons within a broad 3D region of the spinal cord. Such a combinatorial method will better serve basic research in neuroscience.
For high-resolution 3D imaging of the neuronal architecture, sample preparation is very important, which will largely influence the image quality. Specific neuron labeling and high contrast between the neuronal site and surrounding background helps to achieve high-resolution 3D imaging of neuronal structure. By far, Golgi staining is still important in neuroscience research due to its whole neuronal labeling characteristic. After years of improvement, the consumption time of Golgi staining has been significantly reduced, and the imaging quality has been increased (Ranjan and Mallick 2010, 2012; Narayanan et al. 2020; Czechowska et al. 2019). However, we find that the GCM still has several shortcomings, such as high background staining, artefacts, and incomplete vasculature labelling (Mizutani et al. 2008; Bentivoglio et al. 2019; Rosoklija et al. 2014). Our study optimized it in spinal cord presentation. First, the current staining frequently results in incomplete vasculature. The residual blood within the tissue will affect the staining, and the neuronal structure will be interfered with the incomplete vascular structure (Mizutani et al. 2008, 2010; Gaballa and Goldman 1999; Monroy-Gomez et al. 2018). However, few studies have mentioned how to improve this issue (Rosoklija et al. 2014). In our study, by infusion of ACSF in the M-GCM, we can effectively diminish the interference of vascular morphology, in contrast to using unperfused fresh specimens for staining in the GCM. Second, background staining and artefact are still an obstacle that reduces the transparency of samples in traditional GCM. During the staining process, residual staining Solution A/B was deposited in the nonneuronal structure areas, which led to inappropriate labeling and opaque tissue background. In current method, we rinsed the tissue in dd-H2O multiple times to elute the residual staining solution and achieved more specific neuron labeling on a clearer background. Last, in the GCM, mounting the tissue sections on the slide at the first step was not good for thoroughly clearing the two sides of tissue sections. By contrast, in our M-GCM method, washing the tissue in dd-H2O was very helpful in reducing the impact of residual staining solution on imaging quality. Methyl salicylate could help to gain high-grade tissue transparency (Senatorov 2002; Hu et al. 2014). Compare with previous studies that used the GCM for spinal cord tissue staining, we got a better visualization of neurons (Khaw et al. 2020; Hong et al. 2019). Therefore, the M-GCM could better serve morphological research of the neuronal network in the future.
In previous studies, 3D imaging of neurons has been achieved through the combination of Golgi staining and advanced microscopes such as two-photon or confocal microscopy (Kassem et al. 2018; Mancuso et al. 2013). However, the field of vision and the tissue penetration of the microscopy was limited, which severely restricts its use for a large sample. SRμCT is a promising and powerful 3D imaging tool that can help us achieve 3D imaging of larger specimens at the micron level. To achieve 3D imaging of neurons in the mass of the spinal cord, the combination of SRμCT and M-GCM was used in the present study. After many tries, high-resolution 3D imaging of neurons with detailed morphological features could be achieved when the distance between the detector and the sample was 3.5 cm and transmissivity of light was 30–70%. Our results showed that M-GCM significantly increases the refractive index of the spinal cord tissue and reduced the artefacts and the incomplete vascular structure on the 3D images. Compared with previous studies, the current method achieved more detailed 3D imaging of neuronal architecture in the mouse spinal cord (Fratini et al. 2015; Bukreeva et al. 2017; Cedola et al. 2017). Furthermore, we achieved a quantitative assessment of the neuronal network, including soma density and axon length. No X-ray radiation damage to cellular morphology was observed in our study. In recent years, a study reported that they achieved 3D imaging of the neuronal architecture of the mouse brain through a combination of SRμCT and Golgi staining (Fonseca et al. 2018). However, their sample preparation procedure of brain tissue was different from the present study. In addition, this combinatorial method has not been used in 3D imaging of neuronal networks in the spinal cord. With our method applied in the research of neuronal morphology, it would allow us to assess the neuronal network in large structures, but do not require tissue sectioning. Accordingly, the new technique presented here will play an essential role in understanding normal and pathological neural networks and quantifying their characteristics.
Conclusion
In conclusion, we modified Golgi-Cox impregnations for better visualization of the neurons and proved that the combination of SRμCT and M-GCM is a powerful method for 3D imaging of detailed neuronal architecture in the mass of the spinal cord.
Data availability
The datasets generated and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- 2D:
-
Two-dimensional
- 3D:
-
Three-dimensional
- SRμCT:
-
Synchrotron radiation micro-computed tomography
- M-GCM:
-
Optimized Golgi Cox method
- GCM:
-
Golgi-Cox method
- dd-H2O:
-
Double-distilled water
- ACSF:
-
Artificial cerebrospinal fluid
References
Bentivoglio M, Cotrufo T, Ferrari S, Tesoriero C, Mariotto S, Bertini G, Berzero A, Mazzarello P (2019) The original histological slides of camillo golgi and his discoveries on neuronal structure. Front Neuroanat 13:3. https://doi.org/10.3389/fnana.2019.00003
Bukreeva I, Campi G, Fratini M, Spano R, Bucci D, Battaglia G, Giove F, Bravin A, Uccelli A, Venturi C, Mastrogiacomo M, Cedola A (2017) Quantitative 3D investigation of Neuronal network in mouse spinal cord model. Sci Rep 7:41054. https://doi.org/10.1038/srep41054
Cao Y, Wu T, Yuan Z, Li D, Ni S, Hu J, Lu H (2015) Three-dimensional imaging of microvasculature in the rat spinal cord following injury. Sci Rep 5:12643. https://doi.org/10.1038/srep12643
Cao Y, Zhou Y, Ni S, Wu T, Li P, Liao S, Hu J, Lu H (2017) Three dimensional quantification of microarchitecture and vessel regeneration by synchrotron radiation microcomputed tomography in a rat model of spinal cord injury. J Neurotrauma 34(6):1187–1199. https://doi.org/10.1089/neu.2016.4697
Cedola A, Bravin A, Bukreeva I, Fratini M, Pacureanu A, Mittone A, Massimi L, Cloetens P, Coan P, Campi G, Spano R, Brun F, Grigoryev V, Petrosino V, Venturi C, Mastrogiacomo M, Kerlero de Rosbo N, Uccelli A (2017) X-ray phase contrast tomography reveals early vascular alterations and neuronal loss in a multiple sclerosis model. Sci Rep 7(1):5890. https://doi.org/10.1038/s41598-017-06251-7
Chen RC, Dreossi D, Mancini L, Menk R, Rigon L, Xiao TQ, Longo R (2012) PITRE: software for phase-sensitive X-ray image processing and tomography reconstruction. J Synchrotron Radiat 19(Pt 5):836–845. https://doi.org/10.1107/S0909049512029731
Czechowska N, van Rienen A, Lang F, Eiberger B, Baader SL (2019) An update on the Golgi staining technique improving cerebellar cell type specificity. Histochem Cell Biol 151(4):327–341. https://doi.org/10.1007/s00418-018-01766-0
Du F (2019) Golgi-Cox staining of neuronal dendrites and dendritic spines with FD rapid GolgiStain Kit. Curr Protoc Neurosci 88(1):e69. https://doi.org/10.1002/cpns.69
Fonseca MC, Araujo BHS, Dias CSB, Archilha NL, Neto DPA, Cavalheiro E, Westfahl H Jr, da Silva AJR, Franchini KG (2018) High-resolution synchrotron-based X-ray microtomography as a tool to unveil the three-dimensional neuronal architecture of the brain. Sci Rep 8(1):12074. https://doi.org/10.1038/s41598-018-30501-x
Fratini M, Bukreeva I, Campi G, Brun F, Tromba G, Modregger P, Bucci D, Battaglia G, Spano R, Mastrogiacomo M, Requardt H, Giove F, Bravin A, Cedola A (2015) Simultaneous submicrometric 3D imaging of the micro-vascular network and the neuronal system in a mouse spinal cord. Sci Rep 5:8514. https://doi.org/10.1038/srep08514
Gaballa MA, Goldman S (1999) Overexpression of endothelium nitric oxide synthase reverses the diminished vasorelaxation in the hindlimb vasculature in ischemic heart failure in vivo. J Mol Cell Cardiol 31(6):1243–1252. https://doi.org/10.1006/jmcc.1999.0956
Hogstrom LJ, Guo SM, Murugadoss K, Bathe M (2016) Advancing multiscale structural mapping of the brain through fluorescence imaging and analysis across length scales. Interface Focus 6(1):20150081. https://doi.org/10.1098/rsfs.2015.0081
Hong Y, Jin Y, Park K, Choi J, Kang H, Lee SR, Hong Y (2019) Elevated serum melatonin under constant darkness enhances neural repair in spinal cord injury through regulation of circadian clock proteins expression. J Clin Med. https://doi.org/10.3390/jcm8020135
Hu J, Cao Y, Wu T, Li D, Lu H (2014) High-resolution three-dimensional visualization of the rat spinal cord microvasculature by synchrotron radiation micro-CT. Med Phys 41(10):101904. https://doi.org/10.1118/1.4894704
Hu J, Cao Y, Wu T, Li D, Lu H (2015) 3D angioarchitecture changes after spinal cord injury in rats using synchrotron radiation phase-contrast tomography. Spinal Cord 53(8):585–590. https://doi.org/10.1038/sc.2015.49
Ian E, Zhao XC, Lande A, Berg BG (2016) Individual neurons confined to distinct antennal-lobe tracts in the heliothine moth: morphological characteristics and global projection patterns. Front Neuroanat 10:101. https://doi.org/10.3389/fnana.2016.00101
Kassem MS, Fok SYY, Smith KL, Kuligowski M, Balleine BW (2018) A novel, modernized Golgi-Cox stain optimized for CLARITY cleared tissue. J Neurosci Methods 294:102–110. https://doi.org/10.1016/j.jneumeth.2017.11.010
Khaw YM, Cunningham C, Tierney A, Sivaguru M, Inoue M (2020) Neutrophil-selective deletion of Cxcr2 protects against CNS neurodegeneration in a mouse model of multiple sclerosis. J Neuroinflammation 17(1):49. https://doi.org/10.1186/s12974-020-1730-y
Lovinger DM (2008) Communication networks in the brain: neurons, receptors, neurotransmitters, and alcohol. Alcohol Res Health 31(3):196–214
Mancuso JJ, Chen Y, Li X, Xue Z, Wong ST (2013) Methods of dendritic spine detection: from Golgi to high-resolution optical imaging. Neuroscience 251:129–140. https://doi.org/10.1016/j.neuroscience.2012.04.010
Mizutani R, Takeuchi A, Uesugi K, Ohyama M, Takekoshi S, Osamura RY, Suzuki Y (2008) Three-dimensional microtomographic imaging of human brain cortex. Brain Res 1199:53–61. https://doi.org/10.1016/j.brainres.2008.01.029
Mizutani R, Takeuchi A, Uesugi K, Takekoshi S, Osamura RY, Suzuki Y (2010) Microtomographic analysis of neuronal circuits of human brain. Cereb Cortex 20(7):1739–1748. https://doi.org/10.1093/cercor/bhp237
Monroy-Gomez J, Santamaria G, Torres-Fernandez O (2018) Overexpression of MAP2 and NF-H associated with dendritic pathology in the spinal cord of mice infected with rabies virus. Viruses. https://doi.org/10.3390/v10030112
Narayanan SN, Bairy LK, Srinivasamurthy SK (2020) Determining factors for optimal neuronal and glial Golgi-Cox staining. Histochem Cell Biol 154(4):431–448. https://doi.org/10.1007/s00418-020-01891-9
Parekh R, Ascoli GA (2013) Neuronal morphology goes digital: a research hub for cellular and system neuroscience. Neuron 77(6):1017–1038. https://doi.org/10.1016/j.neuron.2013.03.008
Ranjan A, Mallick BN (2010) A modified method for consistent and reliable Golgi-cox staining in significantly reduced time. Front Neurol 1:157. https://doi.org/10.3389/fneur.2010.00157
Ranjan A, Mallick BN (2012) Differential staining of glia and neurons by modified Golgi-Cox method. J Neurosci Methods 209(2):269–279. https://doi.org/10.1016/j.jneumeth.2012.06.023
Rosoklija GB, Petrushevski VM, Stankov A, Dika A, Jakovski Z, Pavlovski G, Davcheva N, Lipkin R, Schnieder T, Scobie K, Duma A, Dwork AJ (2014) Reliable and durable Golgi staining of brain tissue from human autopsies and experimental animals. J Neurosci Methods 230:20–29. https://doi.org/10.1016/j.jneumeth.2014.04.006
Saccomano M, Albers J, Tromba G, Dobrivojevic Radmilovic M, Gajovic S, Alves F, Dullin C (2018) Synchrotron inline phase contrast microCT enables detailed virtual histology of embedded soft-tissue samples with and without staining. J Synchrotron Radiat 25(Pt 4):1153–1161. https://doi.org/10.1107/S1600577518005489
Senatorov VV (2002) Dark-field microscopy visualization of unstained axonal pathways using oil of wintergreen. J Neurosci Methods 113(1):59–62. https://doi.org/10.1016/s0165-0270(01)00474-5
St Croix CM, Shand SH, Watkins SC (2005) Confocal microscopy: comparisons, applications, and problems. Biotechniques 39(6 Suppl):S2-5. https://doi.org/10.2144/000112089
Strotton MC, Bodey AJ, Wanelik K, Darrow MC, Medina E, Hobbs C, Rau C, Bradbury EJ (2018) Optimising complementary soft tissue synchrotron X-ray microtomography for reversibly-stained central nervous system samples. Sci Rep 8(1):12017. https://doi.org/10.1038/s41598-018-30520-8
Tesarova M, Heude E, Comai G, Zikmund T, Kaucka M, Adameyko I, Tajbakhsh S, Kaiser J (2019) An interactive and intuitive visualisation method for X-ray computed tomography data of biological samples in 3D Portable Document Format. Sci Rep 9(1):14896. https://doi.org/10.1038/s41598-019-51180-2
Tsai PS, Friedman B, Ifarraguerri AI, Thompson BD, Lev-Ram V, Schaffer CB, Xiong Q, Tsien RY, Squier JA, Kleinfeld D (2003) All-optical histology using ultrashort laser pulses. Neuron 39(1):27–41. https://doi.org/10.1016/s0896-6273(03)00370-2
Wu T, Cao Y, Ni S, Luo Z, Jiang L, Lu H, Hu J (2018) Morphometric analysis of rat spinal cord angioarchitecture by phase contrast radiography: from 2D to 3D Visualization. Spine 43(9):E504–E511. https://doi.org/10.1097/BRS.0000000000002408 (Phila Pa 1976)
Xu Z, Cork LC, Griffin JW, Cleveland DW (1993) Increased expression of neurofilament subunit NF-L produces morphological alterations that resemble the pathology of human motor neuron disease. Cell 73(1):23–33. https://doi.org/10.1016/0092-8674(93)90157-l
Yang P, Gandahi JA, Zhang Q, Zhang LL, Bian XG, Wu L, Liu Y, Chen QS (2013) Quantitative changes of nitrergic neurons during postnatal development of chicken myenteric plexus. J Zhejiang Univ Sci B 14(10):886–895. https://doi.org/10.1631/jzus.B1300005
Acknowledgements
The authors would like to thank Dr. Tiqiao Xiao and other staff at the (BL13W1) station of the Shanghai Synchrotron Radiation Facility (SSRF), Shanghai, China, for their kind assistance during the experiments.
Funding
This research was supported by the National Natural Science Foundation of China (NO.81371956 81874004, 81672174, 81730068), the Hunan Provincial Department of Finance (NO. (2018)82), the Fundamental Research Funds for the Central Universities of Central South University (No.2018zzts254), and the Key R & D Program of the Hunan Provincial Science & Technology Department (NO. 2017SK2061).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Ethics approval
All research protocols were approved by the Animal Ethics Committee of Central South University. Animal care and use were conducted under the guidelines of the Administration Committee of Affairs Concerning Experimental Animals in Hunan Province, China.
Consent for publication
All the authors gave final approval to the version submitted for publication.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Jiang, L., Cao, Y., Yin, X. et al. A combinatorial method to visualize the neuronal network in the mouse spinal cord: combination of a modified Golgi-Cox method and synchrotron radiation micro-computed tomography. Histochem Cell Biol 155, 477–489 (2021). https://doi.org/10.1007/s00418-020-01949-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00418-020-01949-8