Abstract
Purpose
To assess the performance of artificial intelligence in the automated classification of images taken with a tablet device of patients with blepharoptosis and subjects with normal eyelid.
Methods
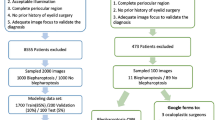
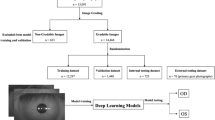
This is a prospective and observational study. A total of 1276 eyelid images (624 images from 347 blepharoptosis cases and 652 images from 367 normal controls) from 606 participants were analyzed. In order to obtain a sufficient number of images for analysis, 1 to 4 eyelid images were obtained from each participant. We developed a model by fully retraining the pre-trained MobileNetV2 convolutional neural network. Subsequently, we verified whether the automatic diagnosis of blepharoptosis was possible using the images. In addition, we visualized how the model captured the features of the test data with Score-CAM. k-fold cross-validation (k = 5) was adopted for splitting the training and validation. Sensitivity, specificity, and the area under the curve (AUC) of the receiver operating characteristic curve for detecting blepharoptosis were examined.
Results
We found the model had a sensitivity of 83.0% (95% confidence interval [CI], 79.8–85.9) and a specificity of 82.5% (95% CI, 79.4–85.4). The accuracy of the validation data was 82.8%, and the AUC was 0.900 (95% CI, 0.882–0.917).
Conclusion
Artificial intelligence was able to classify with high accuracy images of blepharoptosis and normal eyelids taken using a tablet device. Thus, the diagnosis of blepharoptosis with a tablet device is possible at a high level of accuracy.
Trial registration
Date of registration: 2021–06-25.
Trial registration number: UMIN000044660.
Registration site: https://upload.umin.ac.jp/cgi-open-bin/ctr/ctr_view.cgi?recptno=R000051004





Similar content being viewed by others
Data availability
The data that support the findings of this study are available from the corresponding author (DN) upon reasonable request.
Code availability
Not applicable.
References
Koka K, Patel BC (2021) Ptosis correction. In: StatPearls [Internet]. Treasure Island (FL): StatPearls Publishing. https://www.ncbi.nlm.nih.gov/books/NBK539828/. Accessed 21 July 2021
Yadegari S (2016) Approach to a patient with blepharoptosis. Neurol Sci 37:1589–1596. https://doi.org/10.1007/s10072-016-2633-7
Kokubo K, Katori N, Hayashi K, Sugawara J, Fujii A, Maegawa J (2017) Evaluation of the eyebrow position after levator resection. J Plast Reconstr Aesthet Surg 70:85–90. https://doi.org/10.1016/j.bjps.2016.09.025
Zheng X, Kakizaki H, Goto T, Shiraishi A (2016) Digital analysis of eyelid features and eyebrow position following CO2 laser-assisted blepharoptosis surgery. Plast Reconstr Surg Glob Open 4:e1063. https://doi.org/10.1097/GOX.0000000000001063
Hung JY, Perera C, Chen KW et al (2021) A deep learning approach to identify blepharoptosis by convolutional neural networks. Int J Med Inform 148:104402. https://doi.org/10.1016/j.ijmedinf.2021.104402
LeCun Y, Bengio Y, Hinton G (2015) Deep learning. Nature 521:436–444. https://doi.org/10.1038/nature14539
Poplin R, Varadarajan AV, Blumer K et al (2018) Prediction of cardiovascular risk factors from retinal fundus photographs via deep learning. Nat Biomed Eng 2:158–164. https://doi.org/10.1038/s41551-018-0195-0
Gulshan V, Peng L, Coram M et al (2016) Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316:2402–2410. https://doi.org/10.1001/jama.2016.17216
Nagasato D, Tabuchi H, Masumoto H et al (2020) Prediction of age and brachial-ankle pulse-wave velocity using ultra-wide-field pseudo-color images by deep learning. Sci Rep 10:19369. https://doi.org/10.1038/s41598-020-76513-4
Ohsugi H, Tabuchi H, Enno H, Ishitobi N (2017) Accuracy of deep learning, a machine-learning technology, using ultra–wide-field fundus ophthalmoscopy for detecting rhegmatogenous retinal detachment. Sci Rep 7:9425. https://doi.org/10.1038/s41598-017-09891-x
Matsuba S, Tabuchi H, Ohsugi H et al (2019) Accuracy of ultra-wide-field fundus ophthalmoscopy-assisted deep learning, a machine-learning technology, for detecting age-related macular degeneration. Int Ophthalmol 39:1269–1275. https://doi.org/10.1007/s10792-018-0940-0
Masumoto H, Tabuchi H, Nakakura S, Ishitobi N, Miki M, Enno H (2018) Deep-learning classifier with an ultrawide-field scanning laser ophthalmoscope detects glaucoma visual field severity. J Glaucoma 27:647–652. https://doi.org/10.1097/IJG.0000000000000988
Sonobe T, Tabuchi H, Ohsugi H et al (2019) Comparison between support vector machine and deep learning, machine-learning technologies for detecting epiretinal membrane using 3D-OCT. Int Ophthalmol 39:1871–1877. https://doi.org/10.1007/s10792-018-1016-x
Deng J, Dong, W, Socher R, Li L, Kai L, Li F-F (2009) ImageNet: a large-scale hierarchical image database. 2009 IEEE Conference on Computer Vision and Pattern Recognition, pp 248–255.https://doi.org/10.1109/CVPR.2009.5206848
Russakovsky O, Deng J, Su H et al (2015) ImageNet large scale visual recognition challenge. Int J Comp Vision 115:211–252. https://doi.org/10.1007/s11263-015-0816-y
Lee CY, Xie S, Gallagher P, Zhang Z, Tu Z (2015) Deeply-supervised nets. In: Proceedings of the 18th International Conference on Artificial Intelligence and Statistics (AISTATS). San Diego, CA, USA: Journal of Machine Learning Research Workshop and Conference Proceedings, pp 562–570
Mosteller F, Tukey JW (1968) Data analysis, including statistics. In: Lindzey G, Aronson E, eds. Handbook of social psychology: Vol. 2. Research methods. Addison-Wesley, Reading, pp 80–203
Kohavi R (1995) A study of cross-validation and bootstrap for accuracy estimation and model selection. Proc Int Joint Conf AI 2:1137–1145
Sandler M, Howard A, Zhu M, Zhmoginov A, Chen L-C (2018) MobileNetV2: inverted residuals and linear bottlenecks. The IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 4510–4520. arXiv:1801.04381, last revised 21 Mar 2019
Krizhevsky A, Sutskever I, Hinton GE (2012) ImageNet classification with deep convolutional neural networks. Adv Neural Inf Process Syst 1:1097–1105
Agrawal P, Girshick R, Malik J (2014) Analyzing the performance of multilayer neural networks for object recognition. In: Proc Lecture Notes in Computer Science, pp 329–344
Wang H, Wang Z, Du M et al. (2020) Score-CAM: score-weighted visual explanations for convolutional neural networks. Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops 24–25. arXiv:1910.01279, last revised 13 Apr 2020
Liu X, Faes L, Kale AU et al (2019) A comparison of deep learning performance against health-care professionals in detecting diseases from medical imaging: a systematic review and meta-analysis. Lancet Digit Health 1:e271-297. https://doi.org/10.1016/S2589-7500(19)30123-2
Hanley JA, McNeil BJ (1982) The meaning and use of the area under a receiver operating characteristic (ROC) curve. Radiology 143:29–36. https://doi.org/10.1148/radiology.143.1.7063747
Clopper CJ, Pearson ES (1934) The use of confidence or fiducial limits illustrated in the case of the binomial. Biometrika 26:404–413. https://doi.org/10.2307/2331986
Zhang Z, Beck MW, Winkler DA et al (2018) Opening the black box of neural networks: methods for interpreting neural network models in clinical applications. Ann Transl Med 6:216. https://doi.org/10.21037/atm.2018.05.32
Acknowledgements
Yusuke Endo and the orthoptists of Tsukazaki Hospital contributed to the collation of the data. We would like to thank Enago (www.enago.jp) for the English language review.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation was performed by Yoshie Shimizu. Data collection was performed by Naofumi Ishitobi and Hiroki Ochi. Analyses were performed by Hiroki Masumoto and Mao Tanabe. The first draft of the manuscript was written by Hitoshi Tabuchi and Daisuke Nagasato and all authors commented on previous versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval
All images and data obtained and procedures performed in this study were approved by the Institutional Review Board of Tsukazaki Hospital and adhere to the Declaration of Helsinki and its later amendments or comparable ethical standards.
Consent to participate
Written informed consent was obtained from all participants included in the study.
Consent for publication
The authors affirm that human research participants provided informed consent for publication of the images in Fig. 1.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tabuchi, H., Nagasato, D., Masumoto, H. et al. Developing an iOS application that uses machine learning for the automated diagnosis of blepharoptosis. Graefes Arch Clin Exp Ophthalmol 260, 1329–1335 (2022). https://doi.org/10.1007/s00417-021-05475-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00417-021-05475-8