Abstract
Purpose
To automatically detect and classify geographic atrophy (GA) in fundus autofluorescence (FAF) images using a deep learning algorithm.
Methods
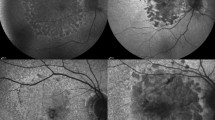
In this study, FAF images of patients with GA, a healthy comparable group and a comparable group with other retinal diseases (ORDs) were used to train a multi-layer deep convolutional neural network (DCNN) (1) to detect GA and (2) to differentiate in GA between a diffuse-trickling pattern (dt-GA) and other GA FAF patterns (ndt-GA) in FAF images.
-
1.
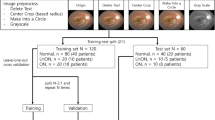
For the automated detection of GA in FAF images, two classifiers were built (GA vs. healthy/GA vs. ORD). The DCNN was trained and validated with 400 FAF images in each case (GA 200, healthy 200, or ORD 200). For the subsequent testing, the built classifiers were then tested with 60 untrained FAF images in each case (AMD 30, healthy 30, or ORD 30). Hereby, both classifiers automatically determined a GA probability score and a normal FAF probability score or an ORD probability score.
-
2.
To automatically differentiate between dt-GA and ndt-GA, the DCNN was trained and validated with 200 FAF images (dt-GA 72; ndt-GA 138). Afterwards, the built classifier was tested with 20 untrained FAF images (dt-GA 10; ndt-GA 10) and a dt-GA probability score and an ndt-GA probability score was calculated.
For both classifiers, the performance of the training and validation procedure after 500 training steps was measured by determining training accuracy, validation accuracy, and cross entropy.
Results
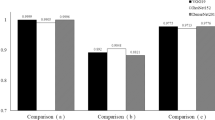
For the GA classifiers (GA vs. healthy/GA vs. ORD), the achieved training accuracy was 99/98%, the validation accuracy 96/91%, and the cross entropy 0.062/0.100. For the dt-GA classifier, the training accuracy was 99%, the validation accuracy 77%, and the cross entropy 0.166.
The mean GA probability score was 0.981 ± 0.048 (GA vs. healthy)/0.972 ± 0.439 (GA vs. ORD) in the GA image group and 0.01 ± 0.016 (healthy)/0.061 ± 0.072 (ORD) in the comparison groups (p < 0.001). The mean dt-GA probability score was 0.807 ± 0.116 in the dt-GA image group and 0.180 ± 0.100 in the ndt-GA image group (p < 0.001).
Conclusion
For the first time, this study describes the use of a deep learning-based algorithm to automatically detect and classify GA in FAF. Hereby, the created classifiers showed excellent results. With further developments, this model may be a tool to predict the individual progression risk of GA and give relevant information for future therapeutic approaches.





Similar content being viewed by others
References
Smith W, Assink J, Klein R, Mitchell P, Klaver C, Klein B, Hofman A, Jensen S, Wang J, de Jong P (2001) Risk factors for age-related macular degeneration. Ophthalmology 108:697–704
Herrmann P, Holz FG, Charbel Issa P (2013) Etiology and pathogenesis of age-related macular degeneration. Ophthalmologe 110:377–387
Bindewald A, Schmitz-Valckenberg S, Jorzik J, Dolar-Szczasny J, Sieber H, Keilhauer C, Weinberger A, Dithmar S, Pauleikhoff D, Mansmann U, Wolf S, Holz F (2005) Classification of abnormal fundus autofluorescence patterns in the junctional zone of geographic atrophy in patients with age related macular degeneration. Br J Ophthalmol 89:874–878
Ferris FL 3rd, Wilkinson CP, Bird A, Chakravarthy U, Chew E, Csaky K, Sadda SR, Beckman Initiative for Macular Research Classification C (2013) Clinical classification of age-related macular degeneration. Ophthalmology 120:844–851
Cole E, Ferrara D, Novais E, Louzada R, Waheed N (2016) Clinical trial endpoints for optical coherence tomography angiography in neovascular age-related macular degeneration. Retina 36(Suppl 1):S83–S92
Regatieri CV, Branchini L, Duker JS (2011) The role of spectral-domain OCT in the diagnosis and management of neovascular age-related macular degeneration. Ophthalmic Surg Lasers Imaging 42(Suppl):S56–S66
Khurana RN, Dupas B, Bressler NM (2010) Agreement of time-domain and spectral-domain optical coherence tomography with fluorescein leakage from choroidal neovascularization. Ophthalmology 117:1376–1380
Ly A, Nivison-Smith L, Assaad N, Kalloniatis M (2017) Fundus autofluorescence in age-related macular degeneration. Optom Vis Sci 94:246–259
Yung M, Klufas MA, Sarraf D (2016) Clinical applications of fundus autofluorescence in retinal disease. Int J Retina Vitreous. https://doi.org/10.1186/s40942-016-0035-x
Fleckenstein M, Mitchell P, Freund KB, Sadda S, Holz FG, Brittain C, Henry EC, Ferrara D (2017) The progression of geographic atrophy secondary to age-related macular degeneration. Ophthalmology. https://doi.org/10.1016/j.ophtha.2017.08.038
Batıoğlu F, Gedik Oğuz Y, Demirel S, Ozmert E (2014) Geographic atrophy progression in eyes with age-related macular degeneration: role of fundus autofluorescence patterns, fellow eye and baseline atrophy area. Ophthalmic Res 52:53–59
Angermueller C, Parnamaa T, Parts L, Stegle O (2016) Deep learning for computational biology. Mol Syst Biol 12:878
Feeny AK, Tadarati M, Freund DE, Bressler NM, Burlina P (2015) Automated segmentation of geographic atrophy of the retinal epithelium via random forests in AREDS color fundus images. Comput Biol Med 65:124–136
Treder M, Lauermann JL, Eter N (2017) Automated detection of exudative age-related macular degeneration in spectral domain optical coherence tomography using deep learning. Graefes Arch Clin Exp Ophthalmol 256:259–265
Wang Y, Zhang Y, Yao Z, Zhao R, Zhou F (2016) Machine learning based detection of age-related macular degeneration (AMD) and diabetic macular edema (DME) from optical coherence tomography (OCT) images. Biomed Opt Express 7:4928–4940
Sun Y, Li S, Sun Z (2017) Fully automated macular pathology detection in retina optical coherence tomography images using sparse coding and dictionary learning. J Biomed Opt 22:16012
Burlina P, Pacheco KD, Joshi N, Freund DE, Bressler NM (2017) Comparing humans and deep learning performance for grading AMD: a study in using universal deep features and transfer learning for automated AMD analysis. Comput Biol Med 82:80–86
Venhuizen F, van Ginneken B, van Asten F, van Grinsven M, Fauser S, Hoyng C, Theelen T, Sánchez C (2017) Automated staging of age-related macular degeneration using optical coherence tomography. Invest Ophthalmol Vis Sci 58:2318–2328
Bogunovic H, Montuoro A, Baratsits M, Karantonis M, Waldstein S, Schlanitz F, Schmidt-Erfurth U (2017) Machine learning of the progression of intermediate age-related macular degeneration based on OCT imaging. Invest Ophthalmol Vis Sci 58:BIO141–BIO150
Bogunovic H, Waldstein S, Schlegl T, Langs G, Sadeghipour A, Liu X, Gerendas B, Osborne A, Schmidt-Erfurth U (2017) Prediction of anti-VEGF treatment requirements in neovascular AMD using a machine learning approach. Invest Ophthalmol Vis Sci 58:3240–3248
Burlina P, Joshi N, Pekala M, Pacheco K, Freund D, Bressler N (2017) Automated grading of age-related macular degeneration from color fundus images using deep convolutional neural networks. JAMA Ophthalmol 135:1170–1176
Aslam TM, Zaki HR, Mahmood S, Ali ZC, Ahmad NA, Thorell MR, Balaskas K (2017) Use of a neural net to model the impact of optical coherence tomography abnormalities on vision in age-related macular degeneration. Am J Ophthalmol. https://doi.org/10.1016/j.ajo.2017.10.015
Prahs P, Radeck V, Mayer C, Cvetkov Y, Cvetkova N, Helbig H, Marker D (2018) OCT-based deep learning algorithm for the evaluation of treatment indication with anti-vascular endothelial growth factor medications. Graefes Arch Clin Exp Ophthalmol 256:91–98
Rampasek L, Goldenberg A (2016) TensorFlow: biology’s gateway to deep learning? Cell Syst 2:12–14
Deng J, Dong W, Socher R, Li L-J, Li K, Fei-Fei L (2009) ImageNet- a large-scale hierarchical image database. CVPR 2009—IEEE conference on computer vision and. Pattern Recogn 2009:248–255
Szegedy C, Vanhoucke V, Ioffe S, Shlens J (2016) Rethinking the inception architecture for computer vision. IEEE Conf Comput Vis Pattern Recognit 2016:2818–2826
TensorFlow (2017) http://www.tensorflow.org/tutorials/image_recognition. TensorFlow. Accessed 30 Jan 2018
Google Developers (2017) https://codelabs.developers.google.com/codelabs/tensorflow-for-poets/#0. Google Developers. Accessed 4 July 2017
Holz FG, Bindewald-Wittich A, Fleckenstein M, Dreyhaupt J, Scholl HP, Schmitz-Valckenberg S, Group FA-S (2007) Progression of geographic atrophy and impact of fundus autofluorescence patterns in age-related macular degeneration. Am J Ophthalmol 143:463–472
Biarnes M, Mones J, Trindade F, Alonso J, Arias L (2012) Intra and interobserver agreement in the classification of fundus autofluorescence patterns in geographic atrophy secondary to age-related macular degeneration. Graefes Arch Clin Exp Ophthalmol 250:485–490
Schmitz-Valckenberg S, Gobel AP, Saur SC, Steinberg JS, Thiele S, Wojek C, Russmann C, Holz FG, For The Modiamd-Study G (2016) Automated retinal image analysis for evaluation of focal hyperpigmentary changes in intermediate age-related macular degeneration. Transl Vis Sci Technol 5:3
Bearelly S, Chau FY, Koreishi A, Stinnett SS, Izatt JA, Toth CA (2009) Spectral domain optical coherence tomography imaging of geographic atrophy margins. Ophthalmology 116:1762–1769
Holz FG, Jorzik J, Schutt F, Flach U, Unnebrink K (2003) Agreement among ophthalmologists in evaluating fluorescein angiograms in patients with neovascular age-related macular degeneration for photodynamic therapy eligibility (FLAP-study). Ophthalmology 110:400–405
Lakhani P, Sundaram B (2017) Deep learning at chest radiography: automated classification of pulmonary tuberculosis by using convolutional neural networks. Radiology 284:574–582
Holz F, Bellman C, Staudt S, Schütt F, Völcker H (2001) Fundus autofluorescence and development of geographic atrophy in age-related macular degeneration. Invest Ophthalmol Vis Sci 42:1051–1056
Schmitz-Valckenberg S, Bindewald-Wittich A, Dolar-Szczasny J, Dreyhaupt J, Wolf S, Scholl HP, Holz FG, Fundus Autofluorescence in Age-Related Macular Degeneration Study G (2006) Correlation between the area of increased autofluorescence surrounding geographic atrophy and disease progression in patients with AMD. Invest Ophthalmol Vis Sci 47:2648–2654
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All authors certify that they have no affiliations with/or involvement in any organization or entity with any financial interest (such as honoraria; educational grants; participation in speakers’ bureaus; membership, employment, consultancies, stock ownership, or other equity interest; and expert testimony or patent-licensing arrangements) or non-financial interest (such as personal or professional relationships, affiliations, knowledge or beliefs) in the subject matter or materials discussed in this manuscript.
Financial disclosure
M. Treder: Allergan, Novartis; J.L. Lauermann: Bayer, Novartis; N. Eter: Allergan, Alimera, Bausch and Lomb, Bayer, Heidelberg Engineering, Novartis, Roche.
Ethical approval
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards.
For this type of study formal consent is not required.
Rights and permissions
About this article
Cite this article
Treder, M., Lauermann, J.L. & Eter, N. Deep learning-based detection and classification of geographic atrophy using a deep convolutional neural network classifier. Graefes Arch Clin Exp Ophthalmol 256, 2053–2060 (2018). https://doi.org/10.1007/s00417-018-4098-2
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00417-018-4098-2