Abstract
Objectives
CLCN4 variations have recently been identified as a genetic cause of X-linked neurodevelopmental disorders. This study aims to broaden the phenotypic spectrum of CLCN4-related condition and correlate it with functional consequences of CLCN4 variants.
Methods
We described 13 individuals with CLCN4-related neurodevelopmental disorder. We analyzed the functional consequence of the unreported variants using heterologous expression, biochemistry, confocal fluorescent microscopy, patch-clamp electrophysiology, and minigene splicing assay.
Results
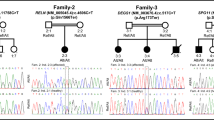
We identified five novel (p.R41W, p.L348V, p.G480R, p.R603W, c.1576 + 5G > A) and three known (p.T203I, p.V275M, p.A555V) pathogenic CLCN4 variants in 13 Chinese patients. The p.V275M variant is found at high frequency and seen in four unrelated individuals. All had global developmental delay (GDD)/intellectual disability (ID). Seizures were present in eight individuals, and 62.5% of them developed refractory epilepsy. Five individuals without seizures showed moderate to severe GDD/ID. Developmental delay precedes seizure onset in most patients. The variants p.R41W, p.L348V, and p.R603W compromise the anion/exchange function of ClC-4. p.R41W partially impairs ClC-3/ClC-4 association. p.G480R reduces ClC-4 expression levels and impairs the heterodimerization with ClC-3. The c.1576 + 5G > A variant causes 22 bp deletion of exon 10.
Conclusions
We further define and broaden the clinical and mutational spectrum of CLCN4-related neurodevelopmental conditions. The p.V275M variant may be a potential hotspot CLCN4 variant in Chinese patients. The five novel variants cause loss of function of ClC-4. Transport dysfunction, protein instability, intracellular trafficking defect, or failure of ClC-4 to oligomerize may contribute to the pathophysiological events leading to CLCN4-related neurodevelopmental disorder.






Similar content being viewed by others
Data availability
The anonymized data are available from the corresponding author upon reasonable request.
References
He HL, Guzman RE, Cao DZ, Sierra-Marquez J, Yin F, Fahlke C et al (2021) The molecular and phenotypic spectrum of CLCN4-related epilepsy. Epilepsia 62:1401–1415
Hu H, Haas SA, Chelly J, Van Esch H, Raynaud M, de Brouwer APM et al (2016) X-exome sequencing of 405 unresolved families identifies seven novel intellectual disability genes. Mol Psychiatry 21:133–148
Palmer EE, Stuhlmann T, Weinert S, Haan E, Van Esch H, Holvoet M et al (2018) De novo and inherited mutations in the X-linked gene CLCN4 are associated with syndromic intellectual disability and behavior and seizure disorders in males and females. Mol Psychiatry 23:222–230
Veeramah KR, Johnstone L, Karafet TM, Wolf D, Sprissler R, Salogiannis J et al (2013) Exome sequencing reveals new causal mutations in children with epileptic encephalopathies. Epilepsia 54:1270–1281
Xu X, Lu F, Zhang L, Li HY, Du SJ, Tang J (2021) Novel CLCN4 variant associated with syndromic X-linked intellectual disability in a Chinese girl: a case report. BMC Pediatr 21:1
Palmer EE, Pusch M, Picollo A, Forwood C, Nguyen MH, Suckow V et al (2023) Functional and clinical studies reveal pathophysiological complexity of CLCN4-related neurodevelopmental condition. Mol Psychiatry 28:668–697
Li SA, Zhang WX, Liang P, Zhu M, Zheng BX, Zhou W et al (2023) Novel variants in the CLCN4 gene associated with syndromic X-linked intellectual disability. Front Neurol 14:1
Guzman RE, Sierra-Marquez J, Bungert-Pluemke S, Franzen A, Fahlke C (2022) Functional characterization of CLCN4 variants associated with X-linked intellectual disability and epilepsy. Front Mol Neurosci 15:1
Rickheit G, Wartosch L, Schaffer S, Stobrawa SM, Novarino G, Weinert S et al (2010) Role of ClC-5 in renal endocytosis is unique among ClC exchangers and does not require PY-motif-dependent Ubiquitylation. J Biol Chem 285:17595–17603
Weinert S, Gimber N, Deuschel D, Stuhlmann T, Puchkov D, Farsi Z et al (2020) Uncoupling endosomal CLC chloride/proton exchange causes severe neurodegeneration. Embo J 39:1
Hur J, Jeong HJ, Park J, Jeon S (2013) Chloride channel 4 is required for nerve growth factor-induced TrkA signaling and neurite outgrowth in PC12 cells and cortical neurons. Neuroscience 253:389–397
Scheel O, Zdebik AA, Lourdel S, Jentsch TJ (2005) Voltage-dependent electrogenic chloride/proton exchange by endosomal CLC proteins. Nature 436:424–427
van Slegtenhorst MA, Bassi MT, Borsani G, Wapenaar MC, Ferrero GB, de Conciliis L et al (1994) A gene from the Xp22.3 region shares homology with voltage-gated chloride channels. Hum Mol Genet 3:547–552
Stauber T, Weinert S, Jentsch TJ (2012) Cell biology and physiology of CLC chloride channels and transporters. Compr Physiol 2:1701–1744
Novarino G, Weinert S, Rickheit G, Jentsch TJ (2010) Endosomal chloride-proton exchange rather than chloride conductance is crucial for renal endocytosis. Science 328:1398–1401
Alekov AK, Fahlke C (2009) Channel-like slippage modes in the human anion/proton exchanger ClC-4. J Gen Physiol 133:485–496
Pusch M, Zifarelli G (2015) ClC-5: physiological role and biophysical mechanisms. Cell Calc 58:57–66
Guzman RE, Grieschat M, Fahlke C, Alekov AK (2013) ClC-3 is an intracellular chloride/proton exchanger with large voltage-dependent nonlinear capacitance. ACS Chem Neurosci 4:994–1003
Smith AJ, Lippiat JD (2010) Direct endosomal acidification by the outwardly rectifying CLC-5 Cl(−)/H(+) exchanger. J Physiol 588:2033–2045
Rohrbough J, Nguyen HN, Lamb FS (2018) Modulation of ClC-3 gating and proton/anion exchange by internal and external protons and the anion selectivity filter. J Physiol 596:4091–4119
Mohammad-Panah R, Harrison R, Dhani S, Ackerley C, Huan LJ, Wang Y et al (2003) The chloride channel ClC-4 contributes to endosomal acidification and trafficking. J Biol Chem 278:29267–29277
Guzman RE, Bungert-Plumke S, Franzen A, Fahlke C (2017) Preferential association with ClC-3 permits sorting of ClC-4 into endosomal compartments. J Biol Chem 292:19055–19065
Jentsch TJ, Pusch M (2018) CLC chloride channels and transporters: structure, function, physiology, and disease. Physiol Rev 98:1493–1590
Duncan AR, Polovitskaya MM, Gaitan-Penas H, Bertelli S, VanNoy GE, Grant PE et al (2021) Unique variants in CLCN3, encoding an endosomal anion/proton exchanger, underlie a spectrum of neurodevelopmental disorders. Am J Hum Genet 108:1450–1465
Guzman RE, Miranda-Laferte E, Franzen A, Fahlke C (2015) Neuronal ClC-3 splice variants differ in subcellular localizations, but mediate identical transport functions. J Biol Chem 290:25851–25862
Laemmli U (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685
Nicke A, Bäumert H, Rettinger J, Eichele A, Lambrecht G, Mutschler E et al (1998) P2X1 and P2X3 receptors form stable trimers: a novel structural motif of ligand-gated ion channels. EMBO J 17:3016–3028
Wittig I, Karas M, Schägger H (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6:1215–1225
Schneider C, Rasband W, Eliceiri K (2012) NIH Image to ImageJ: 25 years of image analysis. Nat Methods 9:671–675
Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232
Zhang W, Ye FH, Chen SM, Peng J, Pang N, Yin F (2022) Splicing interruption by intron variants in causes poirier-bienvenu neurodevelopmental syndrome: a focus on genotype-phenotype correlations. Front Neurosci Switz 16:1
Wittig I, Karas M, Schagger H (2007) High resolution clear native electrophoresis for in-gel functional assays and fluorescence studies of membrane protein complexes. Mol Cell Proteomics 6:1215–1225
Nothmann D, Leinenweber A, Torres-Salazar D, Kovermann P, Hotzy J, Gameiro A et al (2011) Hetero-oligomerization of neuronal glutamate transporters. J Biol Chem 286:3935–3943
Okkenhaug H, Weylandt KH, Carmena D, Wells DJ, Higgins CF, Sardini A (2006) The human ClC-4 protein, a member of the CLC chloride channel/transporter family, is localized to the endoplasmic reticulum by its N-terminus. FASEB J 20:2390–2392
Acknowledgements
We thank the patients and their parents for kindly giving us permission to publish this data. We also thank Tingting Yang and other staff at Beijing Chigene Translational Medicine Research Center for their expert technical assistance. We are grateful to Dr. Daniel Kortzak for generating the ClC-4 structure (Fig. 1a).
Funding
This work was funded by the National Natural Science Foundation of China (82071462, 81771409 to PJ and 82201316 to HH), the Natural Science Foundation of Hunan Province (2022JJ40785 to HH), the China Postdoctoral Science Foundation (2023M733950 to HH), and the Key R&D Program of Hunan Province (2022SK2036 to PJ), the German Research Foundation (DFG) (GU 2042/2-1 to REG).
Author information
Authors and Affiliations
Contributions
Conceptualization: Hailan He, Jing Peng, and Raul E. Guzman; investigation: Hailan He, Raul E. Guzman, Guzman G. A., Stefanie Bungert-Plümke, and Arne Franzen; resources: Hongmin Zhu, Hongwei Zhang, yonglin Yu, Suzhen Sun, Guilan Peng, Zheng Chen, Zhongqin Huang and Jing Peng; collecting and analyzing data: Hailan He, Raul E. Guzman, XueQin Lin, Guzman G. A., Stefanie Bungert-Plümke, and Arne Franzen; writing—original draft preparation: Hailan He; writing—review and editing: Raul E. Guzman, and Jing Peng.
Corresponding authors
Ethics declarations
Conflicts of interest
The authors declare no conflicts of interest.
Ethics approval
This study was conducted following the Declaration of Helsinki and approved by the ethics committee of Xiangya Hospital of Central South University and relevant local ethics committee. Written informed consent was obtained from their parents.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
He, H., Li, X., Guzman, G.A. et al. Expanding the genetic and phenotypic relevance of CLCN4 variants in neurodevelopmental condition: 13 new patients. J Neurol (2024). https://doi.org/10.1007/s00415-024-12383-4
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00415-024-12383-4