Abstract
Background
Charcot–Marie–Tooth disease (CMT) is a genetically and clinically heterogeneous group of inherited neuropathies. Monoallelic pathogenic variants in ATP1A1 were associated with axonal and intermediate CMT. ATP1A1 encodes for the catalytic α1 subunit of the Na+/ K+ ATPase. Besides neuropathy, other associated phenotypes are spastic paraplegia, intellectual disability, and renal hypomagnesemia. We hereby report the first demyelinating CMT case due to a novel ATP1A1 variant.
Methods
Whole-exome sequencing on the patient’s genomic DNA and Sanger sequencing to validate and confirm the segregation of the identified p.P600R ATP1A1 variation were performed. To evaluate functional effects, blood-derived mRNA and protein levels of ATP1A1 and the auxiliary β1 subunit encoded by ATP1B1 were investigated. The ouabain-survival assay was performed in transfected HEK cells to assess cell viability, and two-electrode voltage clamp studies were performed in Xenopus oocytes.
Results
The variant was absent in the local and global control datasets, falls within a highly conserved protein position, and is in a missense-constrained region. The expression levels of ATP1A1 and ATP1B1 were significantly reduced in the patient compared to healthy controls. Electrophysiology indicated that ATP1A1p.P600R injected Xenopus oocytes have reduced Na+/ K+ ATPase function. Moreover, HEK cells transfected with a construct encoding ATP1A1p.P600R harbouring variants that confers ouabain insensitivity displayed a significant decrease in cell viability after ouabain treatment compared to the wild type, further supporting the pathogenicity of this variant.
Conclusion
Our results further confirm the causative role of ATP1A1 in peripheral neuropathy and broaden the mutational and phenotypic spectrum of ATP1A1-associated CMT.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Charcot–Marie–Tooth (CMT) disease is a group of hereditary motor and sensory neuropathies (HMSN) characterized by high genetic and phenotypic heterogeneity [1]. CMT is the most common inherited neuropathy, with a reported global incidence of 1 in 2500 people [2]. CMT is classified into demyelinating (CMT1), axonal (CMT2) or intermediate (CMTI) types based on the neurophysiological criteria. To date, variants in over 120 genes have been linked to CMT pathology.
Pathogenic variants in ATP1A1 (ATPase Na+/K+ Transporting Subunit Alpha 1) were previously associated with peripheral neuropathy. The ATP1A1 gene encodes for the catalytic α1-subunit of the Na+/ K+ ATPase that actively transports the Na+ and K+ ions against their concentration gradients across the cell membrane using the energy of ATP hydrolysis. The pump activity is critical for maintaining resting membrane potential, cell excitability, uptake of nutrients, amino acids, and neurotransmitters, and for maintaining cell volume and intracellular pH. There are four α-subunit isoforms (α1–α4) of the Na+/K+ ATPase encoded by distinct genes that share sequence similarity of more than 75% [3]. ATP1A1 protein is expressed ubiquitously, including in the neurons and glial cells of the brain. Mammals express three auxiliary β subunits (β1–β3). ATP1B1 encoded β1 subunit is expressed almost in all tissues and cells whereas β2 and β3 have restricted tissue expression patterns [4,5,6,7,8]. The β subunit is required to target the Na+/K+ ATPase to the plasma membrane. It also acts as an intracellular adhesion protein by interacting with other β subunits of neighbouring cells [9]. A functional pump is formed by combining one α-, one β-, and optionally one of seven regulatory FXYD subunits [10]. α1 and β1 are the most common heterodimer constituents considered to form the housekeeping Na+/ K+ ATPase isozymes [11].
Variants in ATP1A1, but no other α paralogs were associated with CMT disease to date. Missense variants were identified in ATP1A1 (p.L48R, p.I592T, p.A597T, p.P600T, p.P600A, p.D601F, and p.D811A) in distinct pedigrees affected with autosomal dominant CMT2 [12]. Moreover, two missense variants at positions p.S207F and p.G877S were identified in two Chinese families, and p.G549R was identified in a Spanish proband with intermediate CMT [13, 14].
We identified a novel variant in ATP1A1 that substitutes proline at position 600 for arginine (p.P600R) in a demyelinating CMT patient of Cypriot origin. The variant maps to the previously identified hotspot region of pathogenic axonal CMT variants in ATP1A1 [12]. In this study, we investigated the patient and control blood expression levels of α1 and β1. Furthermore, electrophysiological properties of mutant ATP1A1 versus wild type ATP1A1 were investigated in Xenopus oocytes. Finally, a sodium pump-specific cell viability assay was performed in HEK cells to further characterize the deleterious effects of this novel CMT1-related ATP1A1 variant.
Materials and methods
Patient
Clinical and neurophysiological data were collected from the patient through detailed medical evaluation. Peripheral blood was collected for molecular analysis from the patient and participating controls, including the patient’s healthy brother.
Genetic analysis
Screening of the common CMT1 and CMT4 genes
DNA extraction from whole blood and then targeted gene analysis were performed. Briefly, the duplication of PMP22 was evaluated by Multiple Ligation-Dependent Probe Amplification (MLPA) using the SALSA MLPA kit (MRC-Holland, Amsterdam, Netherlands) according to the manufacturer’s instructions. The results were analysed on the Coffalyser software (MRC-Holland). For Sanger sequencing, primers were designed using the Primer3 software to amplify the coding regions of Cx32, PMP22, MPZ, EGR2, NEFL and GDAP1 by standard PCR. Purified PCR products were sequenced with the BigDye Terminator V.1.1 Cycle Sequencing Kit (ABI, California, USA) and run on a 16-capillary ABI 3130XL sequencer. Sequencing analysis was performed using the SeqScape® Software (ABI, California, USA) by aligning the sample sequence against the reference sequence obtained from the Ensembl genome browser database, human genome assembly: GRCh38p.13 (https://www.ensembl.org/index.html).
Whole-exome sequencing
Whole-exome sequencing (WES) was performed on genomic DNA obtained from the patient’s peripheral blood as previously described [15]. The DNA library was prepared using the Illumina’s TruSeq DNA Exome Kit. Sequencing was performed on the NextSeq-500 platform using the NextSeq 550 High Output kit (150 cycles) according to the manufacturer protocol (Illumina, California, USA). Raw sequence reads were aligned to the human genome reference sequence hg19 using FastQC 0.11.7 and BWA (Aligner) 0.7.15. Imported variants were annotated, filtered, and classified using VariantStudio (Illumina, California, USA). The variants were further filtered using the dbSNP (https://www.ncbi.nlm.nih.gov/snp/) and the Genome Aggregation Database (https://gnomad.broadinstitute.org/). Sanger sequencing was performed to confirm the presence of the NGS-derived variant in the patient. Familial segregation analysis by Sanger sequencing was limited to the patient’s non-affected brother due to the unavailability of parents or other family members.
In silico predictions
A total of five publicly available in silico tools were utilised for predicting variant pathogenicity and conservation of the substituted amino acid, including the Mutation taster (http://www.mutationtaster.org/), PROVEAN (http://provean.jcvi.org/index.php), PolyPhen-2 (http://genetics.bwh.harvard.edu/pph2/), PANTHER (http://www.pantherdb.org/tools/), CADD (https://cadd.gs.washington.edu/snv).
qPCR analysis of gene expression in blood
The total RNA was extracted from the peripheral blood obtained from the patient and healthy controls using the Nucleospin RNA Blood kit (Macherey–Nagel, Duren, Germany). Reverse transcription was performed using the Protoscript cDNA Synthesis Kit (NEB, Massachusetts, USA) according to the manufacturer’s instructions. The starting quantity of 2.5 ng cDNA was determined by the standard curve assay performed on the Quant Studio 7 Flex System (ABI, California, USA). Primers specific to ATP1A1 and ATP1B1 were used, and normalisation was achieved with housekeeping genes GAPDH and β-Actin (Supplementary Table 1). Primers were designed using primer3 to span exon–exon junctions to prevent amplification from the genomic DNA. Primers’ specificities were confirmed using the Primer-Blast software tool (https://www.ncbi.nlm.nih.gov/tools/primer-blast/). The samples were analysed in triplicates in each run, and three independent experiments were performed.
Sequencing of cDNA derived from blood RNA
We investigated whether the observed reduction of ATP1A1 gene expression was due to the absence of the mutant ATP1A1 transcript in the patient blood. The total RNA extracted from patient blood was converted to cDNA. 200 ng cDNA template was amplified by PCR using a set of ATP1A1-specific primers (Forward: ATP1A1_Exon_6_7F; Reverse: ATP1A1_Exon_14R). Agarose gel electrophoresis was performed to verify the correct amplicon size of the PCR product and the product was sequenced with the primers used for PCR amplification. Sequencing analysis was performed on SeqScape software as described above.
Cell culture
Lymphoblastoid cell line (LCLs) cultures
LCLs were chosen for functional analysis since ATP1A1 expression is not detectable in fibroblasts. Patient and control lymphocytes were collected from peripheral blood using Ficoll–Paque Plus upon obtaining written informed consent (Sigma-Aldrich, USA). Selected lymphocytes were infected with the Epstein–Barr virus (EBV) and were cultured in RPMI 1640 medium (Biosera, Nuaille, France), supplemented with 10% FBS (ThermoFisher, Massachusetts, United States), and 50 U/ml of Penicillin/Streptomycin (Biosera) as previously described [16].
Protein extraction
Total protein was isolated from lymphoblastoid cells by cell lysis (50 mM Tris–Cl pH 7.5, 150 mM KCl, 2 mM EDTA, 0.5% Triton x-100 and 1 × protease inhibitor cocktail) followed by one-hour incubation on ice. Cell lysates were sonicated and centrifuged at 4 °C. Protein concentration was determined using the Coomassie Plus (Bradford) protein assay (ThermoFisher Scientific, UK).
Immunoblotting
Ten μg of protein from each lymphoblastoid cell lysate was treated with SDS in the presence of 10% β-mercaptoethanol for denaturation and loaded onto an 8–10% SDS-PAGE gel for separation on a Mini-Protean system (Bio-Rad) and ran at 90 mV for ~ 20 min, then 120 mV until desired migration was achieved. Proteins were transferred onto the Immobilon P-polyvinylidene difluoride (PVDF) membrane by electroblot at a constant 100 mV for 1 h. After the transfer was complete, the PVDF membrane was blocked in PBS containing 0.1% v/v Tween-20 and 5% BSA for one hour. The membranes were incubated overnight at 4 °C with the respective primary antibody. This was followed by three 5–15 min washes in PBS with 0.1% v/v Tween-20. Then, the membranes were incubated with the appropriate horseradish-peroxidase conjugated secondary antibody for 1–2 h at room temperature. Membranes were washed three times in PBS with 0.1% Tween-20 and then incubated with Clarity Western ECL Substrate (Bio-Rad, USA). Visualization of protein bands was processed in the Vilber Fusion Solo X imaging system. Protein expression levels were quantified using the ImageJ software.
Antibodies
Primary antibodies of mouse anti-Na+/K+-ATPase A1/C464.6: sc-21712, Santa-Cruz (1:200), mouse anti-Na+/K+-ATPase β1 (C464.8): sc-21713, Santa-Cruz (1:200), mouse anti-GAPDH/sc-32233, Santa-Cruz (1:10,000) and mouse Anti-β-Actin, Clone AC-74 (A2228), Sigma (1:20,000) were used.
Secondary antibodies of donkey anti-mouse IgG (H + L) SA1-100/Invitrogen (1:10,000) and donkey anti-rabbit IgG (H + L) 31,458/Invitrogen (1:10,000) were used.
Site-directed mutagenesis and heterologous expression in oocytes
P600R was introduced by PCR mutagenesis into the full-length cDNA encoding the ouabain-resistant human α1-isoform (with the mutations Q118R and N129D that mimic the naturally ouabain-resistant rodent α1 isoforms) [17]. For simplicity we refer to the “RD” mutant template as wild type. The resulting plasmid was sequenced, linearized via restriction enzyme (NdeI; New England Biolabs), and transcribed (SP6 Transcription Kit; Thermo Fisher) for injection into Xenopus laevis oocytes. Ovarian follicles (Xenoocyte) were enzymatically digested as previously described [18]. The cRNA of either P600R α1 or wild-type α1 was mixed with cRNA for the β1-isoform at an equimolar ratio and injected into oocytes (75 ng α, 25 ng β). Oocytes were stored at 16 °C in SOS solution (100 mM NaCl, 1 mM MgCl2, 2 mM KCl, 1.8 mM CaCl2, 5 mM HEPES, 2.5 mM pyruvic acid (Sigma-Aldrich), 1 × antibiotic–antimycotic (Gibco), and 5% horse serum (Gibco), titrated to pH 7.5 with NaOH) for 3–7 days.
Electrophysiology
To increase the rate-limiting intracellular Na+ concentration oocytes were incubated in Na+-loading solution (in mM, 90 NaOH, 20 TEA-OH, 40 HEPES, and 0.2 EGTA) with 10 µM ouabain one hour before the two-electrode voltage clamp (TEVC) experiments. Such ouabain concentration inhibited endogenous pumps for the duration of the experiments leaving exogenous pumps unaffected [19].
TEVC was performed with an OC-725C amplifier (Warner Instruments) controlled by pClamp software and a Digidata 1440A (Molecular Devices). Current signals were acquired at 10 kHz and also continuously recorded at 1 kHz via a Minidigi 1B (Molecular Devices). The current and voltage glass microelectrodes were filled with 3 M KCl and had resistances of 0.2–1 MΩ. Oocytes were clamped at a holding potential of − 50 mV from which square voltage steps were applied as indicated.
Experiments were carried out at room temperature 19–21 °C in an extracellular solution containing 122 mM methanesulfonic acid (MS), 5 mM Ba(OH)2, 1 mM Mg(OH)2, 0.5 Ca(OH)2, 10 mM HEPES, 125 mM NaOH (titrated to pH 7.6 with MS, osmolality of 250–260 mOsm/kg). K+ was added to the external solution from a 450 mM K-MS stock. Ouabain (Sigma-Aldrich) was directly dissolved at 10 mM in the external solutions to reversibly inhibit the ouabain-resistant exogenously expressed pumps.
Data analysis
Electrophysiological data was analyzed using Clampfit (part of pClamp software; Molecular Devices) and Origin (OriginLab). Na+/K+ pump-mediated transient currents were calculated subtracting the current induced by voltage pulses in the presence of ouabain from the current in the absence of ouabain. Only the current when the currents return to the baseline was analysed by subtracting the baseline current and integrating the transient current to obtain the charge moved during the voltage pulse. The charge–voltage relationship was fitted with the Boltzmann equation:
where the charge moved by hyperpolarizing pulses is Qhyp, the total charge moved is Qtot, the voltage at the centre of the distribution is V1/2, the apparent valence of a charge crossing the whole electric field is zq, the elementary charge is e, the Boltzmann constant is k, the absolute temperature is T (°K). kT/ezq is commonly referred to the slope factor.
The K+ concentration dependence of the current was fitted by a Hill function:
where I0 is the current at 0 K+, IMax is the current at infinite [K+], K0.5 is the half maximal activating [K+], n is the Hill coefficient.
Ouabain survival assay
Cells were transfected with plasmids encoding full-length human ATP1A1 (WT), and an ATP1A1 construct that has been mutated to be ouabain-insensitive (WT-oua). Cells transfected with plasmid constructs to express wild-type (WT-oua) and selected mutants (p.P600R-oua, p.G509D-oua, p.G718S-oua) were treated with 0.5 μM ouabain over 72 h. The p.G509D and p.G718S mutants were included as positive controls, involved in a complex neurodevelopmental syndrome, and were included in this analysis due to their proximity to the p.P600 variant. Cell survival was measured using the CellTiter-Glo luciferase assay.
Statistical analysis
WB: Quantitative data from at least three independent experiments were analysed with one-way ANOVA. P value < 0.05 was considered as statistically significant. Results are expressed as mean% ± SE (Standard error) from the three independent experiments. The mean quantitative data of the controls was normalised to 100%.
OSA: Experiments were conducted in replicates of eight, and survival of a specific mutant was normalized to untreated cells transfected with the same plasmid. Normalized cell survival under ouabain treatment was compared by ANOVA, using the Tukey’s post-test for alpha-error correction.
Results
Patient phenotype
The proband is a 50-year-old female who first presented with progressive leg and hand weakness at the age of 5 years. Nerve conduction studies performed at the age of 12 years showed prolonged ulnar and median motor nerve distal latencies (median: 5.2 ms; ulnar: 5.6 ms) and slowing of motor nerve conduction velocities (MNCVs) between 22.2 m/s (ulnar) and 27.4 m/s (median), with low compound muscle action potential (CMAP) amplitudes (median: 1.9 mV; ulnar: 3.0 mV) showing temporal dispersion. Motor responses in the lower limbs and sensory responses in both upper and lower limbs (median ulnar, sural) were all unobtainable. Median and ulnar F-wave responses were also abolished. Selective needle electromyography showed severely reduced recruitment of high amplitude, long duration, polyphasic motor unit potentials and spontaneous activity present in the tibial anterior, peroneus longus and medial gastrocnemius muscles, consistent with severe chronic-active neurogenic process.
At age 14, she had a tenotomy of the Achilles tendon, while she slowly developed more proximal muscle weakness and started having gait dysfunction after adolescence. In parallel, she developed weakness of the hand muscles, atrophy of all distal muscles and decreasing function in both arms and legs.
At the age of 43, the patient reported having difficulties climbing stairs in recent years and stated that she felt much weaker overall. At last examination, she needed help with all activities of daily living. Her family history is negative for any neuromuscular disease. She had a healthy older brother, and both of her deceased parents were reported to be neurologically unaffected.
The patient had severe bilateral sensorineural hearing loss, weakness and atrophy of all distal limb muscles (1–2/5 MRC scale distal to the elbows and knees), mild-moderate weakness of proximal muscles (4 + upper limbs, 4 to 4− lower limbs), areflexia, and severe sensory loss in a stocking-glove distribution. She walked with an unsteady steppage gait and impaired balance. The CMT neuropathy score (CMTNSv2) was 34.
Electrodiagnostic studies at that time revealed unobtainable motor nerve responses from upper and lower limbs with the exception of radial motor response showing prolonged distal latency (6.1 ms), reduced CMAP amplitude (0.1 mV) and severe slowing of the MNCV (8.0 m/s). Severe chronic denervation was detected by needle EMG in the triceps brachii muscle. Further neurophysiological evaluation with brainstem auditory evoked potentials (BAEP) confirmed severe peripheral hearing loss, while visual evoked responses were normal.
In the absence of family history and based on her neurological and electrodiagnostic findings, the clinical diagnosis was established as demyelinating CMT type 1 or 4.
Identification of the novel ATP1A1 c.1799 C > G missense variant in the CMT1 patient
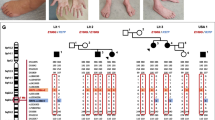
MLPA analysis of PMP22 duplication and Sanger sequencing of the coding regions of the most common CMT genes; Cx32, PMP22, MPZ, EGR2, NEFL, and GDAP1 were negative in the patient. WES has identified a heterozygous missense variant, c.1799C > G (NM_000701.8), in exon 13 of ATP1A1. Sanger sequencing confirmed the variant in the patient, whereas her healthy brother was found to be negative (Fig. 1A). The variant was absent in the large dataset of GnomAD and the in-house dataset (100 WES) of the Cypriot population. The amino acid sequence at the position of the variant was highly conserved, affecting a highly constrained region, and predicted to be damaging according to the in silico prediction tools (Fig. 1B, C). A CADD score of 25.7 further suggested the deleteriousness of this variant. The 67 bp region including the p.600P in ATP1A1 has a constraint coding region (CCR) score of ≥ 99th percentile, indicating that the region is under purifying selection and is highly constrained for genomic variation (https://s3.us-east-2.amazonaws.com/ccrs/ccr.html) [20]. The inheritance pattern of the variant could not be investigated due to the absence of parents who were reported to be neurologically unaffected. In addition, the negative family history for any neuromuscular disorder suggests that the patient could have acquired the variant de novo.
Genetic findings of the Cypriot proband. A Electropherogram of the control ATP1A1 sequence (upper) versus patient ATP1A1: c.1799C > G (p.P600R) sequence (lower). B In silico analysis of the ATP1A1: c.1799C > G (p.P600R). C Conversation analysis of amino acid sequences of ATP1A1 (NM_000701.8) across species compared to p.P600R
RNA and protein expression analysis in the CMT1 patient and controls
ATP1A1 expression
Relative mRNA and protein expression was investigated by performing qPCR analysis and Western blotting. The results of the qPCR analysis indicated a reduction in the expression of ATP1A1 mRNA to approximately 50% in patient blood compared to healthy controls (Fig. 2A). Western blot analysis performed in patient-derived lymphoblasts further confirmed the significant reduction of ATP1A1 protein expression in the CMT1 patient compared to healthy controls (Fig. 2B, C).
mRNA and protein expression analysis of the wild type and mutant ATP1A1. A mRNA expression of ATP1A1 exons in the patient peripheral blood cells compared to healthy controls (Patient: 58.04 ± 2.16%, Control 1: 100.25 ± 2.32%, Control 2: 122.32 ± 6.54%, Control 3: 111.76 ± 2.77%. P = 0.00002). Three independent qPCRs were performed and samples were run as triplicates in each experiment. B Western blot analysis results of ATP1A1 expression normalized against β-ACTIN and GAPDH. C Quantification of protein expression of ATP1A1 in the patient versus healthy control LCLs (Patient: 45.49 ± 10.02%, Control 1: 99.01 ± 14.84%, Control 2: 94.15 ± 9.10%, Control 3: 123.94 ± 17.00%. P = 0.002). WB was replicated eight times for quantification. Statistical analysis was performed by one-way ANOVA. P < 0.05 considered statistically significant. D The expression of mutant ATP1A1 (c.1799 C > G, P600R) transcript in patient blood confirmed by Sanger sequencing
ATP1A1 P600R transcript detection
RNA from patient blood was converted into cDNA and used as a template for ATP1A1 sequencing of the region containing the c.1799C > G change (Fig. 2D). The mutant transcript was expressed in equal level with the wild type transcript in the patient, indicating that the observed reduction of ATP1A1 transcript levels in lymphoblastoid cells is not an mRNA synthesis defect due to the c.1799C > G variant.
ATP1B1 expression
We investigated the ATP1B1 expression levels in the patient blood due to its ubiquitous expression and the most prevalent associate of the α1 subunit amongst other β subunits. The β1 subunit contains N-glycosylated forms in the range of ~ 60–40 kDa consisting of mature, hybrid and immature types [21] (Fig. 3B). Investigation of ATP1B1 mRNA and protein levels were performed by qPCR and Western blot analysis, respectively. ATP1B1 mRNA was reduced to ~ 45% in patient blood in comparison to healthy controls (Fig. 3C). The protein levels of β1 subunit in LCLs were further reduced to ~ 30% in the patient compared to healthy controls.
mRNA and protein expression analysis of ATP1B1. A mRNA expression analysis of ATP1B1 cDNA in the patient peripheral blood cells compared to healthy controls (Patient: 44.71 ± 3.63%, Control 1: 100.04 ± 3.40%, Control 2: 94.63 ± 4.31%, Control 3: 103.99 ± 3.21%. P = 0.00001). Four independent qPCRs were performed and samples were analysed as triplicates in each run. B Western blot analysis results of ATP1B1 expression normalized against β-ACTIN and GAPDH. C) ATP1B1 protein expression quantification in patient versus healthy control LCLs (Patient: 30.22 ± 6.43%, Control 1: 118.20 ± 13.21%, Control 2: 110.05 ± 10.23%, Control 3: 103.64 ± 11.04%. P = 0.0002). WB was replicated four times for quantification. Statistical analysis was performed by one-way ANOVA. P < 0.05 considered statistically significant
Electrophysiology
TEVC was performed in WT (ouabain resistant) versus mutant P600R ATP1A1 injected Xenopus oocytes to determine the electrophysiological characteristics of the pump function (Fig. 4). The concentration dependence of outward current activated by K+ (Fig. 4A) illustrates the reduced maximal pump current (Fig. 4B), without alteration of the K0.5 for K+. The maximal pump current is proportional to both the number of pumps and their turnover rate. A better estimation of the number of functional pumps in the plasma membrane can be obtained by measuring the total charge moved in transient currents elicited by voltage pulses in the absence of K+ (Fig. 4D). As these transient currents report on the conformational change of the pump, without cycling, the total charge moved across the whole voltage range is only proportional to the number of pumps (independent of the turnover rate). Upon integration of the transient currents the mean total charge from Boltzmann fits to individual experiments was Qtot = 21.6 ± 10.6 nC for WT and 15.3 ± 11.1 for P600R (n = 29, SD for both). The Qtot from P600R-injected oocytes normalized to WT-injected oocytes injected and measured on the same days, was reduced by about 50% (Fig. 4E). Effects of a mutation on the apparent affinity for external Na+ can be estimated from the centre of the Boltzmann distribution in the Q–V curves [18, 19] (Fig. 4F). The minor ~ 10 mV rightward shift induced by P600R indicates a 50% increase in affinity for external Na+, that is not expected to be of pathological significance. Thus, the results in Xenopus oocytes suggest that the pathological effects of the P600R mutation are not due to changes in affinities for Na+ or K+.
Electrophysiological findings. A Representative traces from an oocyte expressing WT (that is the RD ouabain resistant version) and P600R pumps. Oocytes were held at − 50 mV and exposed to the indicated [K+] (in mM) to activate pump current. Vertical deflections indicate application of 100 ms-long pulses to measure voltage-dependent parameters. B K0.5 for K+ activation of pump current obtained from Hill fits (“Methods”) to the [K+] dependencies at each voltage, as a function of the applied voltage. C Current, normalized to the mean from oocytes expressing WT pumps on the same day under the same conditions (results from 3 batches of oocytes, 11 WT and 13 P600R oocytes total. D Ouabain-sensitive transient currents elicited by pulses from − 50 mV to voltages ranging from − 140 mV to + 40 mV (in 20 mV increments). E Charge (integral of current transients) as a function of voltage normalized to the total charge moved in the whole voltage range (Qtot = 21.6 ± 10.6 for WT and 15.3 ± 11.1 for P600R). Line plots are Boltzmann distributions fitted to the data (mean V0.5 = − 47.5 ± 4.3 mV for WT and − 38.1 ± 4.3 mV for P600R, n = 29 each). F Mean total charge (measured from Boltzmann distributions in individual experiments) normalized to the mean from oocytes expressing WT pumps on the same day
Functional effect of P600R variant on cell viability
The functional effect of the c.1799 C > G (p.P600R) variant was further evaluated with an ouabain survival assay using HEK cells transfected with ouabain-insensitive ATP1A1 constructs as described [12]. Cell death was observed in the cells transfected with WT ATP1A1, whereas cells transfected with the ouabain-resistant WT (WT-oua) survived. Cells transfected with selected mutants demonstrated a significant decrease in viability in all groups. Cells transfected with p.G509D-oua and p.G718S-oua, two positive controls described in association with a severe neurodevelopmental syndrome, showed a higher cell death proportion than the p.P600R-oua (Fig. 5).
Discussion
We present a Cypriot patient with early-onset, progressive demyelinating CMT accompanied by severe sensorineural hearing loss. WES identified a novel c.1799C > G (p.P600R) missense change in ATP1A1 in this patient. This variant was absent in the general and local population datasets. The ancestral amino acid is highly conserved and its region is highly constrained for missense variants. In silico pathogenicity prediction tools suggested a damaging consequence for the p.P600R variant. The absence of this variant in the healthy brother and the negative family history for any neuromuscular disease suggests that the patient acquired the c.1799C > G variant de novo.
The clinical findings in this case expand the spectrum of previously reported phenotypes associated with ATP1A1 variants. Although intermediate slowing of NCV has been described [12] no MNCVs below 30 m/s were reported. In our patient, upper limb MNCVs were clearly in the demyelinating range, with only moderately reduced CMAP amplitudes in childhood. This may be explained by the fact that in addition to axons, ATP1A1 has been shown to be prominently expressed at the non-compact myelin areas of myelinating Schwann cells including the paranodal loops and Schmidt-Lantermann incisures [12]. Moreover, ATP1A1 and ATP1A3 showed mutually exclusive expression patterns throughout the spinal cord and ATP1A1 was strongly expressed in the myelin sheath of lumbar roots. Thus, it is plausible that dysfunction of this subunit can cause disturbed homeostasis starting in Schwann cells, with concurrent or secondary axonal degeneration, consistent with the phenotype in this case. Similar mechanisms have been described in other CMT genes associated with phenotypes spanning the axonal to intermediate and demyelinating range in terms of electrophysiology, as in CMT1X caused by variants in GJB1/Connexin32 gene also expressed at non-compact myelin [22]. Another novel finding in this case was the sensorineural hearing loss. Although the onset is unclear from the patient’s history because of slow progression over the years, it was clearly documented at the age of 43. Hearing loss of variable severity may occur in patients with both axonal and demyelinating inherited neuropathies [23] and it is possible that it might have been underrecognized in previous reports of patients affected by ATP1A1 variants, if not reported by the patients and no BAEPs have been performed. De novo ATP1A1 variants in CMT are rare and have not been genetically confirmed (current study and ref 12). Previously, de novo ATP1A1 variants were reported in hypomagnesemia, spastic paraplegia, sleep disorder, intellectual disability and epileptic encephalopathy and, complex neurodevelopmental syndrome [24,25,26,27], whereas, inherited ATP1A1 variants were associated with axonal or intermediate CMT [12,13,14] (Table 1). Six out of ten (60%) CMT2-ATP1A1 variants map to the large cytoplasmic loop between transmembrane domains M4 and M5, which contains the ATP binding (N) and the phosphorylation (P) site (Fig. 6). Two of these variants alter proline at position 600 to alanine or threonine. Here, we report a demyelinating CMT case due to a novel hotspot ATP1A1 variant, p.P600R.
ATP1A1 protein and respective locations of the pathogenic variants reported up to date. Variants written in black represent CMT related variants identified in other studies, red represents the ATP1A1 variant identified in this study, blue represent hypomagnesaemia and intellectual disability or spastic paraplegia, green represent developmental delay, orange represent complex neurodevelopmental syndrome
Analysis of ATP1A1 expression revealed a ~ 50% downregulation of its mRNA and protein levels in the CMT patient’s blood compared to healthy controls. To investigate the mechanism of this downregulation, we sequenced the patient derived cDNA spanning the c.1799C > G variant region. Results revealed that the mutant allele was able to produce the transcript in a similar amount to the wild type allele without frameshift or exon-skipping, confirming that the observed reduction of ATP1A1 expression is not due to reduced biosynthesis or a splicing defect of the mutant transcript. The β subunit of the Na+/K+ ATPase is important for stabilizing the α subunit and its export from the ER to the plasma membrane, hence preventing its degradation [21, 28, 29]. The β subunit is also able to modify ion transport properties of the catalytic α subunit thus, modulating the activity of its α counterpart [30,31,32]. We investigated whether the expression level of ATP1B1, the ubiquitously expressed form of β would have been affected due to reduced ATP1A1 levels in our CMT patient. Indeed, we identified a significant downregulation of ATP1B1 mRNA and protein levels in our CMT patient compared to controls, similar to ATP1A1 expression levels. This may suggest a dominant negative effect of the p.P600R variant. Alternatively, another mechanism might be playing a role for the regulation of the Na+/K+ ATPase expression by affecting the transcript levels of its subunits.
Electrophysiology indicated reduced pump function of the p.P600R mutant compared to the ouabain-sensitive WT pump. The mutant had minor effects on pump kinetics in the Xenopus oocytes, which are unlikely to cause the observed loss-of-function, which is probably caused by the reduced expression of the mutant pump seen both in the patient and in the oocyte expression system. Furthermore, ouabain insensitive-p.P600R transfected HEK cells demonstrated a significant decrease in viability compared to wild type ouabain insensitive-ATP1A1 transfected cells, in support of the pathogenicity of this variant. A more substantial cell death was observed in the positive control cells transfected with the p.G509D and p.G718S variants associated with the early-onset complex neurodevelopmental syndrome [27]. The phenotype severity has been reflected in the cell survival ratio as the “milder” p.P600R CMT1 variant caused a relatively less detrimental effect on cell viability compared to clinically more severe positive controls. Pathogenicity of the p.P600 ATP1A1 variants was also shown in axonal/intermediate CMT patients. The p.P600T variant caused a significant decrease in cell viability and the p.P600A variant significantly reduced the Na+-pump currents in Xenopus oocytes, indicating loss of function [12]. Furthermore, the p.P600A dramatically affected the differentiation of patient-derived induced pluripotent stem cells into post-mitotic neurons, suggesting that fewer mature neurons may evolve during neuronal cell development in ATP1A1-CMT [33]. Taken together, alterations of the p.P600 residing at the bridge of the N and P domains of ATP1A1 damage the Na+/K+ ATPase activity and neuronal function.
Previously reported axonal CMT-ATP1A1 variants; p.P600A and p.D811A demonstrated significant reduction in Na+ currents. The p.D811 is a predicted site of Na+ ion coordination suggesting that, missense changes would result in loss of sodium coordination [12]. Since p.P600A resides in the proposed molecular hinge domain, it was suggested that the reduced Na+-pump currents were due to impaired conformational dynamics of the pump during the Na+/K+ transport [12]. Conversely, our electrophysiology analysis on the demyelinating p.P600R variant demonstrated that the reduced pump function was due to reduced expression of the pump proteins instead of a kinetic dysfunction of the Na+/K+ ATPase. In addition, four axonal ATP1A1 variants; p.L48R, p.I592T, p.P600T and p.D811A seem to have more detrimental loss of cell viability (more than two-fold decrease in viability compared to WT-ouabain sensitive cells) in the ouabain survival assay compared to our demyelinating p.P600A variant [12]. This may also reflect the differences in the cellular mechanisms of axonal variants versus the demyelinating p.P600R variant. Furthermore, the two previously identified CMTI mutants; p.S207F and p.G877S were shown to downregulate ATP1A1 protein expression by promoting proteasomal degradation of the mutants without affecting the mRNA levels [13]. Although the main mechanism of pathogenesis seems to be loss of function in all the reported types of ATP1A1-related CMT, the underlying molecular mechanisms for each type clearly have differences, warranting future studies to explore these mechanistic aspects in more detail.
Reduced Na+/K+ ATPase activity at the plasma membrane surface would eventually result in intracellular Na+ build up. The Na+/Ca2+ exchanger responds by reverting to move out excess Na+ while taking Ca2+ into the cytoplasm. This mechanism was previously implicated in various diseases associated with Na+/K+ ATPase mutations [34]. Mitochondria are essential for buffering intracellular Ca2+. Toxic elevation of Ca2+ will eventually lead to mitochondrial dysfunction resulting in increased production of reactive oxygen species (ROS). Mitochondrial ROS stimulates phosphorylation of the α1 subunit by protein kinase PKCζ, which triggers endocytosis of the plasma membrane Na+/K+ ATPase [35,36,37,38]. Phosphorylation-ubiquitination-recognition-endocytosis-degradation (PURED) is an established pathway for regulating plasma membrane proteins including the Na+/K+ ATPase [39,40,41,42]. Phosphorylation of the α1 subunit stimulates ubiquitylation under steady state and hypoxic conditions in alveolar cells and, it inhibits Na+/K+ ATPase in neuronal cells [43,44,45].
Hoxhaj et al., identified that the Thr339-Leu772 fragment of ATP1A1 is a binding site for the closely related E3 ubiquitin ligases; ZNRF1 and ZNRF2 [46]. ZNRF1 and ZNRF2 are expressed in many tissues including in the peripheral nervous system and are targeted to plasma membrane by N-myristoylation [46, 47]. The p.P600R that falls into the binding region of ZNRF1/2 on ATP1A1 might affect the binding of these ubiquitin ligases hence, degradation of the α1 subunit. Altering the degradation dynamics of the tightly controlled Na+/K+ ATPase activity might send a negative feedback message to downregulate the production of α and β transcripts. The ubiquitin–proteasome degradation pathway has been previously linked to the transcriptional regulation of proteins [48,49,50]. Nevertheless, such interplay between degradation and transcriptional regulation of Na+/K+ ATPase subunits needs to be clarified in future studies.
The variability in phenotype due to different amino acid substitutions of the same residue are likely to be due to the nature of the variant. Each substitution may have a different effect on the tertiary structure of the respective protein. Some amino acids would be more damaging due to variability in chemical structures and the bonds they may disrupt or introduce, resulting in altered protein function which may be reflected in the phenotype. For example, the Tyr82Cys variant in MPZ was described in patients with both CMT1 and Dejerine-Sottas Syndrome (DSS) and the Tyr82His change was identified in a milder late-onset phenotype CMT2 patient [51,52,53,54]. The Tyr to His change at position 82 of MPZ protein does not cause a significant structural change [54]. In contrast, introduction of the Cys residue at the same position has a large impact on the tertiary structure of the protein thus, on the myelin structure and function resulting in a more severe phenotype [54]. The ATP1A1 Pro600Arg change hereby presented in a CMT1 patient, likewise may have a different effect on the protein as compared to the Pro600Ala and Pro600Thr variants previously identified in CMT2 patients. Arginine has a larger structure with an electrically charged side-chain whereas, both Threonine and Alanine are smaller and lack an electrically charged side-chain. The wild type residue Proline is smaller than Arginine and has a special structure providing unique properties for the transmembrane proteins, unlikely to be recapitulated by any other amino acid with a different size.
Conclusion
In conclusion, we identified a novel variant at the hotspot CMT residue P600 of ATP1A1 using WES. The proband presented with demyelinating CMT in contrast to the previously identified P600A and P600T variants that cause the axonal type of the disease. Phenotypically, clinical presentation is similar to the previous cases reported [12], although the severity of the neuropathy, in this case, was more pronounced compared to other patients of comparable age group with ATP1A1 variants (CMTNSv2 12–15 in 40–50-year-olds). Furthermore, previously reported upper limb MNCVs in affected patients ranged from 30 to 52 m/s, while our patient presented even lower velocities that were clearly in the demyelinating range. In addition, sensorineural hearing loss found in this patient has not been previously reported in ATP1A1 associated phenotypes. Thus, the phenotypic spectrum of ATP1A1-related CMT appears to expand from the demyelinating to the axonal end of the spectrum, and this should be taken into consideration when investigating demyelinating CMT neuropathies.
Data availability statement
Data are available upon reasonable request.
References
Nicolaou P, Zamba-Papanicolaou E, Koutsou P, Kleopa KA, Georghiou A, Hadjigeorgiou G et al (2010) Charcot–Marie–Tooth disease in cyprus: epidemiological. Clin Genet Charact Neuroepidemiol 35(3):171–177. https://doi.org/10.1159/000314351
Skre H (2008) Genetic and clinical aspects of Charcot–Marie–Tooth’s disease. Clin Genet 6(2):98–118. https://doi.org/10.1111/j.1399-0004.1974.tb00638.x
Shamraj OI, Lingrel JB (1994) A putative fourth Na+, K(+)-ATPase alpha-subunit gene is expressed in testis. Proc Natl Acad Sci 91(26):12952–12956. https://doi.org/10.1073/pnas.91.26.12952
Lavoie L, Levenson R, Martin-Vasallo P, Klip A (1997) The molar ratios of α and β subunits of the Na +−K +-ATPase differ in distinct subcellular membranes from rat skeletal muscle. Biochemistry 36(25):7726–7732. https://doi.org/10.1021/bi970109s
Peng L, Martin-Vasallo P, Sweadner KJ (1997) Isoforms of Na, K-ATPase α and β subunits in the rat cerebellum and in granule cell cultures. J Neurosci 17(10):3488–3502. https://doi.org/10.1523/JNEUROSCI.17-10-03488.1997
Arystarkhova E, Sweadner KJ (1997) Tissue-specific expression of the Na, K-ATPase β3 subunit. J Biol Chem 272(36):22405–22408. https://doi.org/10.1074/jbc.272.36.22405
Martín-Vasallo P, Wetzel RK, García-Segura LM, Molina-Holgado E, Arystarkhova E, Sweadner KJ (2000) Oligodendrocytes in brain and optic nerve express the ?3 subunit isoform of Na, K-ATPase. Glia 31(3):206–218. https://doi.org/10.1002/1098-1136(200009)31:3%3c206::AID-GLIA20%3e3.0.CO;2-1
Wetzel RK, Sweadner KJ (2001) Immunocytochemical localization of NaK-ATPase isoforms in the rat and mouse ocular ciliary epithelium. Investig Ophthalmol Vis Sci 42(3):763–769
Vagin O, Tokhtaeva E, Sachs G (2006) The role of the β1 subunit of the Na, K-ATPase and its glycosylation in cell–cell adhesion. J Biol Chem 281(51):39573–39587. https://doi.org/10.1074/jbc.M606507200
Geering K (2008) Functional roles of Na, K-ATPase subunits. Curr Opin Nephrol Hypertens 17(5):526–532. https://doi.org/10.1097/MNH.0b013e3283036cbf
Blanco G, Mercer RW (1998) Isozymes of the Na-K-ATPase: heterogeneity in structure, diversity in function. Am J Physiol Ren Physiol 275(5):F633–F650. https://doi.org/10.1152/ajprenal.1998.275.5.F633
Lassuthova P, Rebelo AP, Ravenscroft G, Lamont PJ, Davis MR, Manganelli F et al (2018) Mutations in ATP1A1 cause dominant Charcot–Marie–Tooth type 2. Am J Hum Genet 102(3):505–514. https://doi.org/10.1016/j.ajhg.2018.01.023
He J, Guo L, Lin S, Chen W, Xu G, Cai B et al (2019) ATP1A1 mutations cause intermediate Charcot–Marie–Tooth disease. Hum Mutat 40(12):2334–2343. https://doi.org/10.1002/humu.23886
Argente-Escrig H, Frasquet M, Vázquez-Costa JF, Millet-Sancho E, Pitarch I, Tomás-Vila M et al (2021) Pediatric inherited peripheral neuropathy: a prospective study at a Spanish referral center. Ann Clin Transl Neurol 8(9):1809–1816. https://doi.org/10.1002/acn3.51432
Nicolaou P, Tanteles GA, Votsi C, Zamba-Papanicolaou E, Papacostas SS, Christodoulou K, Christou Y-P (2021) A novel CLN6 variant associated with juvenile neuronal ceroid lipofuscinosis in patients with absence of visual loss as a presenting feature. Front Genet. https://doi.org/10.3389/fgene.2021.746101
Nicolaou P, Cianchetti C, Minaidou A, Marrosu G, Zamba-Papanicolaou E, Middleton L, Christodoulou K (2013) A novel LRSAM1 mutation is associated with autosomal dominant axonal Charcot–Marie–Tooth disease. Eur J Hum Genet 21(2):190–194. https://doi.org/10.1038/ejhg.2012.146
Price EM, Lingrel JB (1988) Structure-function relationships in the sodium-potassium ATPase .alpha. subunit: site-directed mutagenesis of glutamine-111 to arginine and asparagine-122 to aspartic acid generates a ouabain-resistant enzyme. Biochemistry 27(22):8400–8408. https://doi.org/10.1021/bi00422a016
Meyer DJ, Gatto C, Artigas P (2017) On the effect of hyperaldosteronism-inducing mutations in Na/K pumps. J Gen Physiol 149(11):1009–1028. https://doi.org/10.1085/jgp.201711827
Spontarelli K, Infield DT, Nielsen HN, Holm R, Young VC, Galpin JD et al (2022) Role of a conserved ion-binding site tyrosine in ion selectivity of the Na+/K+ pump. J Gen Physiol. https://doi.org/10.1085/jgp.202113039
Havrilla JM, Pedersen BS, Layer RM, Quinlan AR (2019) A map of constrained coding regions in the human genome. Nat Genet 51(1):88–95. https://doi.org/10.1038/s41588-018-0294-6
Tokhtaeva E, Sachs G, Vagin O (2009) Assembly with the Na, K-ATPase α 1 subunit is required for export of β 1 and β 2 subunits from the endoplasmic reticulum. Biochemistry 48(48):11421–11431. https://doi.org/10.1021/bi901438z
Vavlitou N, Sargiannidou I, Markoullis K, Kyriacou K, Scherer SS, Kleopa KA (2010) Axonal pathology precedes demyelination in a mouse model of X-linked demyelinating/type I Charcot–Marie tooth neuropathy. J Neuropathol Exp Neurol 69(9):945–958. https://doi.org/10.1097/NEN.0b013e3181efa658
Lerat J, Magdelaine C, Roux A, Darnaud L, Beauvais-Dzugan H, Naud S et al (2019) Hearing loss in inherited peripheral neuropathies: Molecular diagnosis by NGS in a French series. Mol Genet Genom Med. https://doi.org/10.1002/mgg3.839
Schlingmann KP, Bandulik S, Mammen C, Tarailo-Graovac M, Holm R, Baumann M et al (2018) Germline de novo mutations in ATP1A1 cause renal hypomagnesemia, refractory seizures, and intellectual disability. Am J Hum Genet 103(5):808–816. https://doi.org/10.1016/j.ajhg.2018.10.004
Stregapede F, Travaglini L, Rebelo AP, Cintra VP, Bellacchio E, Bosco L et al (2020) Hereditary spastic paraplegia is a novel phenotype for germline de novo ATP1A1 mutation. Clin Genet 97(3):521–526. https://doi.org/10.1111/cge.13668
Lin Z, Li J, Ji T, Wu Y, Gao K, Jiang Y (2021) ATP1A1 de novo mutation-related disorders: clinical and genetic features. Front Pediatr. https://doi.org/10.3389/fped.2021.657256
Dohrn MF, Rebelo AP, Srivastava S, Cappuccio G, Smigiel R, Malhotra et al (2022) De novo ATP1A1 variants in an early-onset complex neurodevelopmental syndrome. Neurology 98(11):440–445. https://doi.org/10.1212/WNL.0000000000013276
Noguchi S, Mishina M, Kawamura M, Numa S (1987) Expression of functional (Na+ + K+)-ATPase from cloned cDNAs. FEBS Lett 225(1–2):27–32. https://doi.org/10.1016/0014-5793(87)81125-0
Fambrough DM, Lemas MV, Hamrick M, Emerick M, Renaud KJ, Inman EM et al (1994) Analysis of subunit assembly of the Na-K-ATPase. Am J Physiol Cell Physiol 266(3):C579–C589. https://doi.org/10.1152/ajpcell.1994.266.3.C579
Eakle KA, Kim KS, Kabalin MA, Farley RA (1992) High-affinity ouabain binding by yeast cells expressing Na+, K(+)-ATPase alpha subunits and the gastric H+, K(+)-ATPase beta subunit. Proc Natl Acad Sci 89(7):2834–2838. https://doi.org/10.1073/pnas.89.7.2834
Jaisser F, Jaunin P, Geering K, Rossier BC, Horisberger JD (1994) Modulation of the Na, K-pump function by beta subunit isoforms. J Gen Physiol 103(4):605–623. https://doi.org/10.1085/jgp.103.4.605
Geering K (2001) The functional role of beta subunits in oligomeric P-type ATPases. J Bioenerg Biomembr 33(5):425–438. https://doi.org/10.1023/a:1010623724749
Manganelli F, Parisi S, Nolano M, Miceli F, Tozza S, Pisciotta C et al (2019) Insights into the pathogenesis of ATP1A1-related CMT disease using patient-specific iPSCs. J Peripher Nerv Syst 24(4):330–339. https://doi.org/10.1111/jns.12357
Biondo ED, Spontarelli K, Ababioh G, Méndez L, Artigas P (2021) Diseases caused by mutations in the Na + /K + pump α1 gene ATP1A1. Am J Physiol Cell Physiol 321(2):C394–C408. https://doi.org/10.1152/ajpcell.00059.2021
Comellas AP, Dada LA, Lecuona E, Pesce LM, Chandel NS, Quesada N et al (2006) Hypoxia-mediated degradation of Na, K-ATPase via mitochondrial reactive oxygen species and the ubiquitin-conjugating system. Circ Res 98(10):1314–1322. https://doi.org/10.1161/01.RES.0000222418.99976.1d
Dada LA, Chandel NS, Ridge KM, Pedemonte C, Bertorello AM, Sznajder JI (2003) Hypoxia-induced endocytosis of Na, K-ATPase in alveolar epithelial cells is mediated by mitochondrial reactive oxygen species and PKC-ζ. J Clin Investig 111(7):1057–1064. https://doi.org/10.1172/JCI16826
Chibalin AV, Ogimoto G, Pedemonte CH, Pressley TA, Katz AI, Féraille E et al (1999) Dopamine-induced endocytosis of Na+, K+-ATPase is initiated by phosphorylation of Ser-18 in the rat α subunit and is responsible for the decreased activity in epithelial cells. J Biol Chem 274(4):1920–1927. https://doi.org/10.1074/jbc.274.4.1920
Chen Z, Krmar RT, Dada L, Efendiev R, Leibiger IB, Pedemonte CH et al (2006) Phosphorylation of adaptor protein–2 μ2 is essential for Na +, K + -ATPase endocytosis in response to either G protein-coupled receptor or reactive oxygen species. Am J Respir Cell Mol Biol 35(1):127–132. https://doi.org/10.1165/rcmb.2006-0044OC
Hicke L (1997) Ubiquitin-dependent internalization and down-regulation of plasma membrane proteins. FASEB J 11(14):1215–1226. https://doi.org/10.1096/fasebj.11.14.9409540
Hicke L, Dunn R (2003) Regulation of membrane protein transport by ubiquitin and ubiquitin-binding proteins. Annu Rev Cell Dev Biol 19(1):141–172. https://doi.org/10.1146/annurev.cellbio.19.110701.154617
Coppi MV, Guidotti G (1997) Ubiquitination of Na, K-ATPase α1 and α2 subunits. FEBS Lett 405(3):281–284. https://doi.org/10.1016/S0014-5793(97)00182-8
Lecuona E, Trejo HE, Sznajder JI (2007) Regulation of Na, K-ATPase during acute lung injury. J Bioenergy Biomembr 39(5–6):391–395. https://doi.org/10.1007/s10863-007-9102-1
Lecuona E, Sun H, Vohwinkel C, Ciechanover A, Sznajder JI (2009) Ubiquitination participates in the lysosomal degradation of Na, K-ATPase in steady-state conditions. Am J Respir Cell Mol Biol 41(6):671–679. https://doi.org/10.1165/rcmb.2008-0365OC
Helenius IT, Dada LA, Sznajder JI (2010) Role of ubiquitination in Na, K-ATPase regulation during lung injury. Proc Am Thorac Soc 7(1):65–70. https://doi.org/10.1513/pats.200907-082JS
Mohan S, Tiwari MN, Biala Y, Yaari Y (2019) Regulation of neuronal Na+/K+-atpase by specific protein kinases and protein phosphatases. J Neurosci 39(28):5440–5451. https://doi.org/10.1523/JNEUROSCI.0265-19.2019
Hoxhaj G, Najafov A, Toth R, Campbell DG, Prescott AR, MacKintosh C (2012) ZNRF2 is released from membranes by growth factors and together with ZNRF1, regulates the Na+/K+ATPase. J Cell Sci 125(19):4662–4675. https://doi.org/10.1242/jcs.110296
Araki T, Milbrandt J (2003) ZNRF proteins constitute a family of presynaptic E3 ubiquitin ligases. J Neurosci 23(28):9385–9394. https://doi.org/10.1523/JNEUROSCI.23-28-09385.2003
Wallace AD, Cidlowski JA (2001) Proteasome-mediated glucocorticoid receptor degradation restricts transcriptional signaling by glucocorticoids. J Biol Chem 276(46):42714–42721. https://doi.org/10.1074/jbc.M106033200
Auld KL, Silver PA (2006) Transcriptional regulation by the proteasome as a mechanism for cellular protein. Cell Cycle 5(14):1503–1505. https://doi.org/10.4161/cc.5.14.2979
Suryadevara V, Willis MS (2019) Walk the line: The role of ubiquitin in regulating transcription in myocytes. Physiology 34(5):327–340. https://doi.org/10.1152/physiol.00055.2018
Himoro M, Yoshikawa H, Matsui T, Mitsui Y, Takahashi M, Kaido M et al (1993) New mutation of the myelin P0 gene in a pedigree of Charcot–Marie–Tooth neuropathy 1. Biochem Mol Biol Int 31(1):169–173
Silander K, Meretoja P, Juvonen V, Ignatius J, Pihko H, Saarinen A, Savontaus M-L (1998) Spectrum of mutations in Finnish patients with Charcot–Marie–Tooth disease and related neuropathies. Hum Mutat 12(1):59–68. https://doi.org/10.1002/(SICI)1098-1004(1998)12:1%3c59::AID-HUMU9%3e3.0.CO;2-A
Boerkoel CF, Takashima H, Garcia CA, Olney RK, Johnson J, Berry K et al (2002) Charcot–Marie–Tooth disease and related neuropathies: Mutation distribution and genotype–phenotype correlation. Ann Neurol 51(2):190–201. https://doi.org/10.1002/ana.10089
Bienfait HME (2006) Late onset axonal Charcot–Marie–Tooth phenotype caused by a novel myelin protein zero mutation. J Neurol Neurosurg Psychiatry 77(4):534–537. https://doi.org/10.1136/jnnp.2005.073437
Acknowledgements
We thank the patient and her brother for their participation in this study.
Funding
This work has been funded by grants from the Cyprus TELETHON (Grant number: 73174), RCB Bank Cyprus and National Science Foundation (Grant number: NSF-MCB 2003251). Functional assessments of variant pathogenicity have been funded by NIH (R01NS105755). MD has received funding by the German Research Foundation (Deutsche Forschungsgemeinschaft, DFG, DO 2386/1-1).
Author information
Authors and Affiliations
Contributions
FCY, PN and KC designed the study. PN and KC coordinated the study. KAK performed clinical evaluation of the proband. PK and AG performed the initial diagnostic tests. FCY and PN performed the genetic and expression analyses. KS and PA performed and analysed electrophysiology studies. MFD, APR and SLZ performed and analysed ouabain survival assay. All authors read, revised and approved the final manuscript.
Corresponding author
Ethics declarations
Conflicts of interest
The authors declare that they have no conflict of interest.
Ethics approval
This study was approved by the National Bioethics Committee of Cyprus (EEBK/EΠ/2013/28).
Informed consent
Written informed consent for genetic analyses and functional experiments of the blood from the participating CMT patient, her healthy brother, and participating control individuals were obtained.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Cinarli Yuksel, F., Nicolaou, P., Spontarelli, K. et al. The phenotypic spectrum of pathogenic ATP1A1 variants expands: the novel p.P600R substitution causes demyelinating Charcot–Marie–Tooth disease. J Neurol 270, 2576–2590 (2023). https://doi.org/10.1007/s00415-023-11581-w
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00415-023-11581-w