Abstract
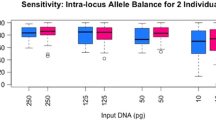
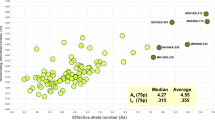
The use of genetic markers, specifically Short Tandem Repeats (STRs), has been a valuable tool for identifying persons of interest. However, the ability to analyze additional markers including Single Nucleotide Polymorphisms (SNPs) and Insertion/Deletion (INDELs) polymorphisms allows laboratories to explore other investigative leads. INDELs were chosen in this study because large panels can be differentiated by size, allowing them to be genotyped by capillary electrophoresis. Moreover, these markers do not produce stutter and are smaller in size than STRs, facilitating the recovery of genetic information from degraded samples. The INDEL Ancestry Informative Markers (AIMs) in this study were selected from the 1000 Genomes Project based on a fixation index (FST) greater than 0.50, high allele frequency divergence, and genetic distance. A total of 25 INDEL-AIMs were optimized and validated according to SWGDAM guidelines in a five-dye multiplex. To validate the panel, genotyping was performed on 155 unrelated individuals from four ancestral groups (Caucasian, African, Hispanic, and East Asian). Bayesian clustering and principal component analysis (PCA) were performed revealing clear separation among three groups, with some observed overlap within the Hispanic group. Additionally, the PCA results were compared against a training set of 793 samples from the 1000 Genomes Project, demonstrating consistent results. Validation studies showed the assay to be reproducible, tolerant to common inhibitors, robust with challenging casework type samples, and sensitive down to 125 pg. In conclusion, our results demonstrated the robustness and effectiveness of a 25 loci INDEL system for ancestry inference of four ancestries commonly found in the United States.





Similar content being viewed by others
Data availability
Authors can confirm that all relevant data are included in the article and/or its supplementary information files.
References
Andersen JF, Greenhalgh MJ, Butler HR, Kilpatrick SR, Piercy RC et al (1996) Further validation of a multiplex STR system for use in routine forensic identity testing. Forensic Sci Int 78:47–64. https://doi.org/10.1016/0379-0738(95)01861-1
Romanini C, Catelli ML, Borosky A, Pereira R, Romero M et al (2012) Typing short amplicon binary polymorphisms: supplementary SNP and Indel genetic information in the analysis of highly degraded skeletal remains. Forensic Sci Int Genet 6:469–476. https://doi.org/10.1016/j.fsigen.2011.10.006
Takahashi M, Kato Y, Mukoyama H, Kanaya H, Kamiyama S (1997) Evaluation of five polymorphic microsatellite markers for typing DNA from decomposed human tissues–correlation between the size of the alleles and that of the template DNA. Forensic Sci Int 90:1–9. https://doi.org/10.1016/s0379-0738(97)00129-1
Dixon LA, Dobbins AE, Pulker HK, Butler JM, Vallone PM et al (2006) Analysis of artificially degraded DNA using STRs and SNPs - results of a collaborative European (EDNAP) exercise. Forensic Sci Int 164:33–44. https://doi.org/10.1016/j.forsciint.2005.11.011
Butler JM, Coble MD, Vallone PM (2007) STRs vs. SNPs: thoughts on the future of forensic DNA testing. Forensic Sci Med Pathol 3:200–205. https://doi.org/10.1007/s12024-007-0018-1
Pereira R, Phillips C, Alves C, Amorim A, Carracedo A et al (2009) A new multiplex for human identification using insertion/deletion polymorphisms. Electrophoresis 30:3682–3690. https://doi.org/10.1002/elps.200900274
Mills RE, Luttig CT, Larkins CE, Beauchamp A, Tsui C et al (2006) An initial map of insertion and deletion (INDEL) variation in the human genome. Genome Res 16:1182–1190. https://doi.org/10.1101/gr.4565806
Mullaney JM, Mills RE, Pittard WS, Devine SE (2010) Small insertions and deletions (INDELs) in human genomes. Hum Mol Genet 19:R131–R136. https://doi.org/10.1093/hmg/ddq400
Kidd JM, Cooper GM, Donahue WF, Hayden HS, Sampas N et al (2008) Mapping and sequencing of structural variation from eight human genomes. Nature 453:56–64. https://doi.org/10.1038/nature06862
LaRue BL, Ge J, King JL, Budowle B (2012) A validation study of the Qiagen Investigator DIPplex(R) kit; an INDEL-based assay for human identification. Int J Legal Med 126:533–540. https://doi.org/10.1007/s00414-012-0667-9
Hollard C, Mendisco F, Keyser C, Crubézy E, Ludes B (2011) First application of the Investigator DIPplex indels typing kit for the analysis of ancient DNA samples. Forensic Sci Int Genet Suppl Ser 3:e393–e4. https://doi.org/10.1016/j.fsigss.2011.09.058
Klein R, Neumann C, Roy R (2015) Detection of insertion/deletion polymorphisms from challenged samples using the Investigator DIPplex(R) kit. Forensic Sci Int Genet 16:29–37. https://doi.org/10.1016/j.fsigen.2014.11.020
Budowle B, van Daal A (2008) Forensically relevant SNP classes. Biotechniques 44:603–608. https://doi.org/10.2144/000112806
Nachman MW, Crowell SL (2000) Estimate of the mutation rate per nucleotide in humans. Genetics 156:297–304. https://doi.org/10.1093/genetics/156.1.297
Zhu Q, Cao Y, Zhang S, Huang Y, Hu Y et al (2021) A targeted ancestry informative InDels panel on capillary electrophoresis for ancestry inference in Asian populations. Electrophoresis 42:1605–1613. https://doi.org/10.1002/elps.202100016
Zhang X, Shen C, Jin X, Guo Y, Xie T et al (2021) Developmental validations of a self-developed 39 AIM-InDel panel and its forensic efficiency evaluations in the Shaanxi Han population. Int J Legal Med 135:1359–1367. https://doi.org/10.1007/s00414-021-02600-4
Inacio A, Costa HA, da Silva CV, Ribeiro T, Porto MJ et al (2017) Study of InDel genetic markers with forensic and ancestry informative interest in PALOP’s immigrant populations in Lisboa. Int J Legal Med 131:657–660. https://doi.org/10.1007/s00414-016-1484-3
Santos C, Phillips C, Oldoni F, Amigo J, Fondevila M et al (2015) Completion of a worldwide reference panel of samples for an ancestry informative indel assay. Forensic Sci Int Genet 17:75–80. https://doi.org/10.1016/j.fsigen.2015.03.011
Holsinger KE, Weir BS (2009) Genetics in geographically structured populations: defining, estimating and interpreting F(ST). Nat Rev Genet 10:639–650. https://doi.org/10.1038/nrg2611
Scientific Working Group on DNA Analysis Methods (2016) Validation Guidelines for DNA Analysis Methods. SWGDAM Documents. [Web] https://www.swgdam.org/_files/ugd/4344b0_813b241e8944497e99b9c45b163b76bd.pdf. Accessed January 2023
QIAGEN (2022) EZ1&2® DNA Investigator® Kit Handbook. [Web] https://www.qiagen.com/us/resources/download.aspx?id=46064856-1b88-4b27-a825-d3f616e06c08&=en. Accessed January 2023
QIAGEN (2016) Investigator® Lyse&Spin Basket Kit Handbook. [Web] https://www.qiagen.com/us/resources/download.aspx?id=3850f41c-fec4-464a-8044-3211268c435b&=en. Accessed January 2023
Thompson LM, Zeng X, Sage K, Sturm S, LaRue B (2015) Selection of an Ancestry-Informative Marker (AIM) Panel of INDELs. Thesis, University of North Texas Health Science Center at Fort Worth
Danecek P, Auton A, Abecasis G, Albers CA, Banks E et al (2011) The variant call format and VCFtools. Bioinformatics 27:2156–2158. https://doi.org/10.1093/bioinformatics/btr330
Ye J, Coulouris G, Zaretskaya I, Cutcutache I, Rozen S et al (2012) Primer-BLAST: a tool to design target-specific primers for polymerase chain reaction. BMC Bioinformatics 13:134. https://doi.org/10.1186/1471-2105-13-134
Shen Z, Qu W, Wang W, Lu Y, Wu Y et al (2010) MPprimer: a program for reliable multiplex PCR primer design. BMC Bioinformatics 11:143. https://doi.org/10.1186/1471-2105-11-143
Sturm SA (2016) A Novel Multiplex Assay for an Ancestry-Informative Marker (AIM) Panel of INDELs. Thesis, University of North Texas
Biosystems A, Hot Start LD (2010) Strong Finish™ Protoc [Web] https://assets.thermofisher.com/TFS-Assets/LSG/manuals/cms_041347.pdf
QIAGEN (2011) QIAGEN® Multiplex PCR Plus Kit. [Web] https://www.qiagen.com/us/resources/download.aspx?id=d30998e1-fbcf-47f3-a415-93c6b9e0863f&=en
ThermoFisher (2019) GlobalFiler™ and GlobalFiler™ IQC PCR Amplification Kits [Web] https://www.thermofisher.com/document-connect/document-connect.html?url=https://assets.thermofisher.com/TFS-Assets%2FLSG%2Fmanuals%2F4477604.pdf
ThermoFisher (2015) DNeasy® Plant Handbook. [Web] https://www.qiagen.com/us/resources/download.aspx?id=f6455f80-dc4f-4ff2-b2de-ae7a3e6c91e0&=en.
Hammer Ø, Harper DAT, Ryan PD (2001) PAST: Paleontological Statistics Software Package for Education and Data Analysis. Palaeontologia Electronica 4:9
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620. https://doi.org/10.1111/j.1365-294X.2005.02553.x
Earl DA, vonHoldt BM (2011) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361. https://doi.org/10.1007/s12686-011-9548-7
Lewis PO, Zaykin D (2001) Genetic Data Analysis: Computer program for the analysis of allelic data. University of Connecticut, Storrs. [Computer Program] http://lewis.eeb.uconn.edu/lewishome/software.html
QIAGEN (2016) Technical Information Investigator® DIPplex Kit. [Web] https://www.qiagen.com/us/resources/download.aspx?id=b6604482-cade-4386-87f0-fa5d0cddcdf4&=en
Elwick K, Mayes C, Hughes-Stamm S (2018) Comparative sensitivity and inhibitor tolerance of GlobalFiler(R) PCR amplification and investigator(R) 24plex QS kits for challenging samples. Leg Med (Tokyo) 32:31–36. https://doi.org/10.1016/j.legalmed.2018.01.006
Huang Y, Liu C, Xiao C, Chen X, Yi S et al (2020) Development of a new 32-plex InDels panel for forensic purpose. Forensic Sci Int Genet 44:102171. https://doi.org/10.1016/j.fsigen.2019.102171
Hara M, Kido A, Kameyama H, Takada A, Miyazaki T et al (2008) STR and Y-STR genotyping assays of 25-year-old semen stains. Forensic Sci Int Genet Suppl Ser 1:428–429. https://doi.org/10.1016/j.fsigss.2007.10.061
Maciejewska A, Jakubowska J, Pawlowski R (2013) Whole genome amplification of degraded and nondegraded DNA for forensic purposes. Int J Legal Med 127:309–319. https://doi.org/10.1007/s00414-012-0764-9
Pereira R, Phillips C, Pinto N, Santos C, dos Santos SE et al (2012) Straightforward inference of ancestry and admixture proportions through ancestry-informative insertion deletion multiplexing. PLoS ONE 7:e29684. https://doi.org/10.1371/journal.pone.0029684
Karlsson S, Mork J (2005) Deviation from hardy–Weinberg equilibrium, and temporal instability in allele frequencies at microsatellite loci in a local population of Atlantic Cod. ICES J Mar Sci 62:1588–1596. https://doi.org/10.1016/j.icesjms.2005.05.009
Chen B, Cole JW, Grond-Ginsbach C (2017) Departure from Hardy Weinberg Equilibrium and genotyping error. Front Genet 8:167. https://doi.org/10.3389/fgene.2017.00167
Bryc K, Durand EY, Macpherson JM, Reich D, Mountain JL (2015) The genetic ancestry of African americans, latinos, and European americans across the United States. Am J Hum Genet 96:37–53. https://doi.org/10.1016/j.ajhg.2014.11.010
Acknowledgements
We would like to acknowledge the Sam Houston State University Department of Forensic Science as well as the Southeast Texas Applied Forensic Science Facility and the donors and their loved ones, without whom this research would not be possible. We would also like to thank Natalia Czado for her helpful edits to this manuscript.
Funding
This study was partially funded by the National Institute of Justice Award #2013-DN-BX-K036. The opinions, findings, conclusions, or recommendations expressed in this presentation are those of the authors and do not necessarily reflect those of the National Institute of Justice.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Compliance with ethical standards
The hair, blood, and buccal samples used in this study were collected in accordance with Institutional Review Board (IRB) approved protocols (IRB-FY2017-32804). All procedures were in accordance with the 1964 Helsinki Declaration and its later amendments.
Conflict of interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.










Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Avellaneda, L.L., Johnson, D.T., Gutierrez, R.M. et al. Development of a novel five dye insertion/deletion (INDEL) panel for ancestry determination. Int J Legal Med (2024). https://doi.org/10.1007/s00414-024-03196-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s00414-024-03196-1