Abstract
In the age of next-generation sequencing (NGS) and with the availability of whole sequenced genomes and epigenomes, some attention has shifted from purely sequence-based studies to those of heritable epigenetic modifications. Transgenerational inheritance can be defined as heritable changes to the state of DNA that may be passed on to subsequent generations without alterations to the underlying DNA sequence. Although this phenomenon has been extensively studied in many systems, studies of transgenerational inheritance in mammals and other higher-level eukaryotes may be complicated by the fact that many epigenetic marks are reprogrammed during sexual reproduction. This, by definition, may obscure our interpretation of what is in fact truly transgenerational. Therefore, in this mini review, we discuss what is currently known in the field about transgenerational epigenetic inheritance in ciliates and plants, with a particular emphasis on RNA-mediated processes and changes in chromatin states.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
In recent years, there has been much focus on the subject of epigenetic inheritance and how heritable changes in chromatin states may be transmitted to subsequent generations. This includes changes in gene activity and gene expression levels, without altering the underlying DNA sequence, that may be passed on to generations to come. Transgenerational inheritance may involve DNA methylation or other chromatin-based mechanisms, but can also involve RNA-mediated DNA methylation and RNA-mediated DNA excision/elimination in some of the more extreme examples. Indeed, small non-coding RNAs have been implicated in many of these processes and likely mediate transgenerational inheritance across eukaryotic species, since they can induce changes in chromatin dynamics and guide histone modifications. While these phenomena have been described in mammals and extensive work has been performed to elucidate mechanisms, this review will focus primarily on RNA-mediated transgenerational inheritance in ciliated protozoans and plants.
Ciliates
Ciliates are large, unicellular protists that can be found ubiquitously across the globe in both marine and freshwater environments. Ciliates exhibit a special case of germline-soma specialization called nuclear dimorphism and are thus unique systems to study RNA-mediated transgenerational inheritance. Ciliates contain two completely separate caches of genetic information: the micronucleus (MIC), considered the germline nucleus, is transcriptionally silent and is used to propagate genetic information from one generation to the next and the macronucleus (MAC), considered the somatic nucleus, is used for vegetative growth of the cells (Prescott 1994). The micronuclear genome resembles that of a canonical eukaryotic genome, with many genes organized along long chromosomes. The micronuclear genome contains a large amount of “junk” DNA including transposable elements (TEs) and repetitive elements such as minisatellites, while micronuclear genes themselves are often interrupted by multiple short transposon-derived stretches of non-coding DNA called internally eliminated sequences (IESs) (Arnaiz et al. 2012; Chen et al. 2014; Guerin et al. 2017; Hamilton et al. 2016). The macronuclear genome, on the other hand, is devoid of all of this “junk” DNA and all the transcription necessary for vegetative growth occurs here (Aury et al. 2006; Duret et al. 2008; Eisen et al. 2006; Fang et al. 2012; Swart et al. 2013). During the ciliate sexual life cycle, the parental macronucleus provides genetic information in the form of transported sRNAs for the formation of a new macronucleus, which is derived from a newly formed, micronuclear precursor. During this micronucleus to macronucleus transition, the micronuclear genome is modified drastically through various processing events, including the polytenization of chromosomes and removal of repetitive DNA sequences, and IESs must be precisely removed to create functional macronuclear genes. It has been shown that small RNAs (sRNAs) are involved in the epigenetic transmission of information from parental nuclei to the developing macronucleus, leading to large-scale genomic rearrangements, altered chromatin states and ultimately the complete removal of specific DNA sequences.
In the stichotrich Oxytricha trifallax, approximately 20% of micronuclear genes exist in a non-linear, “scrambled” order that must be connected upon IES removal during macronuclear development (Chen et al. 2014). This means that in addition to targeting specific IES regions for elimination from the genome, the cells must also sort and reorder the remaining macronuclear destined sequences (MDSs) into functional genes. During Oxytricha conjugation, the parental macronucleus is broken down and degraded, while a new macronucleus, called the anlage, develops from one of the parental micronuclei. At this developmental stage, the anlage undergoes endoreplication, eliminates over 90% of its germline genome, breaks apart and fragments its chromosomes, and then ligates the thousands of remaining MDSs back together into functional genic reading frames (Adl and Berger 2000). A conjugation-specific class of 27 nt small RNAs called 27macRNAs has been identified and is highly upregulated during this process (peaking 24 h post-mixing of complementary mating types) (Fang et al. 2012; Zahler et al. 2012). These 27macRNAs are derived from the parental macronucleus, possess a strong 5′ U bias, and play a vital role during macronuclear development. The 27macRNAs associate with a PIWI protein called Otiwi1 and have been implicated in specifying which regions of the genome are protected from the DNA elimination occurring during this time (Fang et al. 2012). Microinjection of synthetic sRNAs corresponding to IES regions that are usually eliminated led to their retention in subsequent generations. Although little is known about the biogenesis of these RNAs or the exact mechanism by which DNA is protected, it has been suggested that this may occur through methylation of cytosine residues within IES regions (Bracht et al. 2012). In addition to the 27macRNAs necessary for DNA retention throughout macronuclear development, long maternal guide RNA templates transcribed from macronuclear nanochromosomes have also been shown to mediate genomic rearrangements (Nowacki et al. 2008). Long RNA transcripts (both sense and antisense), corresponding to entire macronuclear DNA molecules, can be detected for a brief period during conjugation and it is hypothesized that these act as templates for the correct unscrambling of MDSs. Microinjection of synthetic double-stranded nanochromosomes (DNA or RNA versions) with alternatively arranged MDSs led to defects in the proper reordering of MDSs in subsequent generations, indicating epigenetic inheritance through these RNAs (Nowacki et al. 2008). In a recent study, RNA-cached copies of over half of Oxytricha nanochromosomes have been identified during macronuclear development, supporting the model in which maternal guide RNA templates are transmitted to the progeny (Lindblad et al. 2017). Although it has been suggested that the long guide RNAs may act as precursors for the biogenesis of 27macRNAs, the relationship between these two classes of RNAs remains unknown. Interestingly, a striking number of the genes upregulated during Oxytricha macronuclear development encode well-conserved proteins with links to germline function and development in higher-level eukaryotes (Neeb et al. 2017).
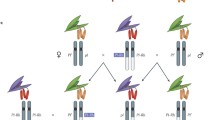
In the more well studied ciliates Paramecium and Tetrahymena, it has also been shown that epigenetic information from the parental macronucleus guides the elimination and subsequent retention of specific DNA sequences during macronuclear development (Fig. 1) (Aronica et al. 2008; Lepere et al. 2008; Mochizuki et al. 2002). During the early stages of the sexual life cycle of these ciliates, the entire micronuclear genome is transcribed bidirectionally to produce long double-stranded RNAs (Chalker and Yao 2001; Mochizuki and Gorovsky 2004b). These double-stranded RNA precursors are then processed by Dicer-like enzymes, DCL2/3 in Paramecium and Dcl1p in Tetrahymena, to produce a class of small RNAs called scan RNAs (scnRNAs) (25 nt and 26–31 nt, respectively) (Chalker et al. 2005; Malone et al. 2005; Mochizuki and Gorovsky 2004a, 2005; Sandoval et al. 2014). These scnRNAs are then transported to the maternal macronucleus where they “scan” the macronuclear genome. Although the mechanism of this genome “scanning” is unknown, it is thought to involve interaction between transported scnRNAs and maternal RNA transcripts present in the developing macronucleus (Aronica et al. 2008; Lepere et al. 2008; Mochizuki et al. 2002). scnRNAs with homologous macronuclear sequence are degraded by an unknown mechanism, leaving only those corresponding to IESs remaining. According to the current model, the scnRNAs that survive this filtering step are transported to the developing macronucleus, where in association with PIWI proteins (Ptiwi1/9 in Paramecium and Twi1p in Tetrahymena) they are hypothesized to mark IESs for excision and elimination (Bouhouche et al. 2011; Duharcourt et al. 1995; Mochizuki et al. 2002; Mochizuki and Gorovsky 2004a; Sandoval et al. 2014). In both Paramecium and Tetrahymena, this elimination relies on repressive heterochromatin marks, namely histone H3 lysine 9 and lysine 27 methylation (Kataoka and Mochizuki 2011; Lhuillier-Akakpo et al. 2014; Liu et al. 2007; Mochizuki and Gorovsky 2004a; Taverna et al. 2002; Yao and Chao 2005). Excision of IES regions is facilitated by a domesticated piggyBac transposase (called PiggyMac in Paramecium), an endonuclease that creates DNA double-stranded breaks at MDS/IES junctions, and flanking MDSs are then joined by the protein DNA ligase IV (Baudry et al. 2009; Cheng et al. 2010; Dubois et al. 2012; Kapusta et al. 2011). In Paramecium, excised IESs circularize to become templates for the transcription of a second class of RNAs called iesRNAs (Allen et al. 2017; Betermier et al. 2000; Kapusta et al. 2011). These 22–31 nt small RNAs, complementary to the sequence of excised IESs, are produced by the Dicer-like enzyme DCL5 and act as a quality control mechanism to ensure the precise and accurate removal of all remaining IESs from developing macronuclear chromosomes (Sandoval et al. 2014). A second class of scnRNAs called late-scnRNAs, expressed later in macronuclear development, has also been reported in Tetrahymena. These late-scnRNAs are transcribed from IESs prior to their excision and can recognize not only the IESs from which they are transcribed, but also other IESs in trans (Noto et al. 2015). The mechanisms by which these lately expressed sRNAs recognize IESs within the developing macronuclear genomes remain to be elucidated; however, Paramecium iesRNAs have been recently shown to bind the previously unclassified PIWI proteins Ptiwi10 and Ptiwi11 (Furrer et al. 2017). It is worth noting that while the Paramecium micronuclear genome contains close to 45,000 IESs, the majority of which interrupt protein-coding regions, Tetrahymena, have significantly less (~ 8000) with very few contained within protein-coding genes (Arnaiz et al. 2012; Fass et al. 2011).
Scanning model for DNA elimination in Paramecium tetraurelia. (a) The micronuclear genome is transcribed bidirectionally by an unknown RNA polymerase to produce long double-stranded RNAs. (b) These long dsRNA precursors are processed by the Dicer-like enzymes DCL2/3 to produce 25 nt long scnRNAs. (c) scnRNAs, in association with the PIWI proteins Ptiwi1/9, are transported to the maternal macronucleus (MAC). (d) scnRNAs “scan” the macronuclear genome via interaction with RNA transcripts of somatic DNA. scnRNAs pairing to homologous macronuclear destined sequences (MDSs) are filtered out and degraded, leaving only those corresponding to germline internal eliminated sequences (IESs). (e) Selected scnRNAs, in association with PIWI proteins Ptiwi10/11, are transported to the developing MAC. (f) These scnRNAs target the excision of IESs by the excisase PiggyMac (PGM). (g) Excised IESs circularize, or concatemerize and circularize, and are transcribed into long dsRNAs. (h) These long dsRNA precursors are processed by the Dicer-like enzyme DCL5 to produce 22–31 nt long iesRNAs. (i) iesRNAs ensure the precise and efficient excision of all remaining IESs from the developing macronuclear genome. Development of the new MAC is completed, with the newly formed macronuclear genome matching that of the maternal macronucleus
Plants
Epigenetic inheritance has been most well studied in plants and generally involves heritable changes in DNA methylation states. Although extensive work has been performed to describe transgenerational silencing of transgenes in plants such as toadflax, tomato, and maize, here we will focus primarily on RNA-directed DNA methylation (RdDM) in the plant Arabidopsis thaliana. Interestingly, compared to mammals, plants are particularly prone to epigenetic inheritance even though both types of genomes tend to be saturated with TEs and other repetitive DNA sequences that must be silenced (Quadrana and Colot 2016). Arabidopsis and other flowering plants exhibit the best characterized example of duplication and functional specification of subunits of the RNA polymerase II complex. In addition to the canonical RNA polymerase II machinery, nearly universally composed of 12 core subunits in eukaryotes, Arabidopsis possesses two additional nuclear multi-subunit RNA polymerases, named RNA polymerase IV (Pol IV) and RNA polymerase IV (Pol V) (reviewed in (Haag and Pikaard 2011) (Kornberg 2007). These plant-specific RNA polymerases have non-redundant roles in RNA-mediated gene silencing pathways, specifically in RNA-directed DNA methylation (RdDM) that targets TEs and other repetitive sequences (Matzke and Mosher 2014; Tucker et al. 2010). Pol IV is responsible for transcribing short primary RNA transcripts, which are copied into dsRNAs by an RNA-dependent RNA polymerase, RDR2 (Blevins et al. 2015; Kasschau et al. 2007; Xie et al. 2004; Zhai et al. 2015; Zhang et al. 2007). After these double-stranded substrates are cleaved by the Dicer-like enzyme DCL3 to produce 24 nt siRNAs, they are stabilized by a 3′ end modification (2′-O-CH3 group) added by the methylase HEN1 (Li et al. 2005; Qi et al. 2005; Xie et al. 2004; Yu et al. 2005). These stabile siRNAs then associate with the Argonaute family protein AGO4 (or sometimes AGO6 and AGO9) to form an RNA-induced silencing complex (RISC) (Blevins et al. 2015; Havecker et al. 2010; Qi et al. 2006; Zhai et al. 2015). Pol V produces nascent long non-coding RNA (lncRNA) transcripts from specified regions of the genome which are hypothesized to base pair with the AGO4-associated siRNAs and this results in de novo cytosine methylation of the corresponding DNA template by the DNA methyltransferase DRM2 (Haag et al. 2009; Wierzbicki et al. 2008; Wierzbicki et al. 2009; Zhong et al. 2014). This often leads to gene silencing through repressive histone modifications (Kanno et al. 2010; Law and Jacobsen 2010) (Fig. 2). Proteomic analyses have revealed that Arabidopsis Pol IV and Pol V have a 12-subunit composition like Pol II. In fact, half of the subunits of Pols II, IV, and V are encoded by the same genes. The remaining Pol IV- or Pol V-specific subunit genes arose through duplication and subfunctionalization of ancestral Pol II subunit genes (Haag and Pikaard 2011). Unique paralogs of the largest subunit of Pol II (NRPB1) make up the catalytic core of the polymerases and are unique to either the Pol IV or Pol V complex, being referred to as NRPD1 and NRPE1, respectively (Herr et al. 2005; Kanno et al. 2005; Onodera et al. 2005; Pontier et al. 2005). While the NRPB1 C-terminal domain (CTD) contains heptapeptide repeats, the CTDs of both NRPD1 and NRPE1 lack this signature, likely facilitating their alternative functions. The NRPE1 CTD is extended by approximately 300 amino acids and is shown to associate with AGO4 through WG/GW repeats, called the Argonaute “hook,” to direct DNA methylation (Li et al. 2006).
RNA-directed DNA methylation (RdDM) in Arabidopsis thaliana. (a) RNA polymerase IV produces single-stranded RNA transcripts that initiate the process of RdDM. (b) These single-stranded RNAs are used as templates for the transcription of a second strand by the RNA-dependent RNA polymerase RDR2. (c) These double-stranded RNA duplexes are cleaved by the Dicer-like enzyme DCL3 to produce 24 nt siRNAs. (d) 24 nt siRNA duplexes are modified by the RNA methylase HEN1, which adds a stabilizing 3′-O-methyl group. (e) A single 24 nt strand of RNA is loaded onto the Argonaute family protein AGO4 to form an active RNA-induced silencing complex (RISC). (f) This sRNA bound RISC complex is then recruited to growing transcripts produced by RNA polymerase V, where direct interaction between AGO4-bound sRNAs and nascent transcripts is thought to occur. (g) De novo cytosine methylation of the corresponding DNA sequence is mediated by the methyltransferase DRM2. This ultimately leads to the removal of active histone marks and the establishment of repressive ones, leading to silencing of specific genomic regions
To be considered truly transgenerational, these DNA methylation landscapes must be heritable and maintained in subsequent generations after their initial establishment. Maintenance of DNA methylation patterns through DNA replication requires the cooperation of several protein factors (Law and Jacobsen 2010). In Arabidopsis, DNA methylation is well maintained across TEs and genes and relies primarily on the de novo DNA methyltransferase (DMTase) MET1 for maintenance of CG methylation (Vongs et al. 1993). For maintenance of CHG methylation, an additional chromomethylase called CMT3 is necessary and specifically binds histone H3 lysine 9 dimethylation, while asymmetric CHH methylation maintenance relies on DRM2 and CMT2, only requiring CMT3 at specific loci (Cao et al. 2003; Du et al. 2015; Johnson et al. 2007; Lindroth et al. 2001; Lindroth et al. 2004; Stroud et al. 2014; Stroud et al. 2013). Evidence suggests that methylation patterns across TEs and repetitive sequence elements are transmitted from parent to offspring upon fertilization, although CHH methylation must be reestablished as the embryo develops (Hsieh et al. 2009; Jullien et al. 2012). CHH methylation is guided by maternally inherited 24 nt siRNAs that are present upon fertilization (Calarco et al. 2012; Lu et al. 2012; Mosher et al. 2009). Plants tend to undergo significantly less germline reprogramming of DNA methylation patterns than mammals, displaying an excellent example of transgenerational epigenetic inheritance (Heard and Martienssen 2014).
Perspectives
Both plants and ciliates exhibit a case of RNA-mediated epigenetic inheritance, utilizing classes of small RNAs, but the mechanisms by which they perform such a feat are quite different. While RNA polymerase IV and RNA polymerase V are involved in the transcription of sRNA precursors and the nascent transcript targets in plants, this process is much less clear in ciliates. Plants use an RNA-dependent RNA polymerase (RDRP) to transcribe the second strand of RNA before Dicer-like cleavage, but it is hypothesized that in ciliates, transcription occurs bidirectionally to form dsRNA substrates, although the polymerase responsible has yet to be identified. Interestingly, however, additional RNA polymerase II subunits have also been identified in ciliates that may play similar roles to plant Pols IV and V. For example, Oxytricha has additional largest and second largest Pol II subunit paralogs (RPB1b and RPB2b) that are highly upregulated during macronuclear development and likely play roles in the transcription of either small RNA precursors or the guide RNAs described in the process of gene unscrambling (Khurana et al. 2014; Neeb et al. 2017). These systems also use two distinct types of Argonaute family RNA-binding proteins to target regions of the genome for silencing or excision, using the AGOs or Piwis, respectively. AGOs are completely absent from ciliate genomes and Piwis have taken on the roles of these proteins. It remains unclear how Piwi-bound small RNAs “target” particular regions of the genome, but one can imagine the involvement of nascent transcripts within the developing macronucleus. Interestingly, plant AGO4 which binds siRNAs is most similar to Ptiwi10, known to bind iesRNAs in Paramecium during the second IES removal step of the “scanning model,” suggesting a possible similar mechanism (Furrer et al. 2017). Potentially, the most striking difference between these two systems is the fact that while both plants and ciliates silence particular regions of the genome using repressive histone modifications, plants merely form heterochromatin, while ciliates like Tetrahymena use repressive histone marks to excise and degrade large segments of the genome entirely, taking this process to the extreme. It remains to be elucidated whether ciliates and plants share additional commonalities in how they accomplish epigenetic inheritance and additional studies are needed to fill in the remaining gaps in our understanding of these models.
Conclusion
Ciliates and plants represent unique and fascinating systems to study RNA-mediated transgenerational epigenetic inheritance. Both genomes must protect against the invasion of transposable elements and other foreign DNA, and this comes in the form of silencing expression, and in some cases excision, of entire DNA sequences to carefully defend the germline and subsequent generations. With the power of next-generation sequencing (NGS) of entire genomes and epigenomes, along with reverse genetic approaches, it will be possible to investigate the roles of epigenetic inheritance in other biological processes and contexts. Although transgenerational inheritance is clearly demonstrated and well described in ciliates and plants, further work is needed to investigate the implications in mammalian systems and how widespread this process is among other eukaryotes.
References
Adl SM, Berger JD (2000) Timing of life cycle morphogenesis in synchronous samples of Sterkiella histriomuscorum. II. The sexual pathway. J Eukaryot Microbiol 47(5):443–449. https://doi.org/10.1111/j.1550-7408.2000.tb00073.x
Allen SE, Hug I, Pabian S, Rzeszutek I, Hoehener C, Nowacki M (2017) Circular concatemers of ultra-short DNA segments produce regulatory RNAs. Cell 168(6):990–999 e997. https://doi.org/10.1016/j.cell.2017.02.020
Arnaiz O, Mathy N, Baudry C, Malinsky S, Aury JM, Denby Wilkes C, Garnier O, Labadie K, Lauderdale BE, Le Mouel A et al (2012) The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet 8(10):e1002984. https://doi.org/10.1371/journal.pgen.1002984
Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K (2008) Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev 22(16):2228–2241. https://doi.org/10.1101/gad.481908
Aury JM, Jaillon O, Duret L, Noel B, Jubin C, Porcel BM, Segurens B, Daubin V, Anthouard V, Aiach N et al (2006) Global trends of whole-genome duplications revealed by the ciliate Paramecium tetraurelia. Nature 444(7116):171–178. https://doi.org/10.1038/nature05230
Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Betermier M (2009) PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev 23(21):2478–2483. https://doi.org/10.1101/gad.547309
Betermier M, Duharcourt S, Seitz H, Meyer E (2000) Timing of developmentally programmed excision and circularization of Paramecium internal eliminated sequences. Mol Cell Biol 20(5):1553–1561. https://doi.org/10.1128/MCB.20.5.1553-1561.2000
Blevins T, Podicheti R, Mishra V, Marasco M, Wang J, Rusch D, Tang H, Pikaard CS (2015) Identification of Pol IV and RDR2-dependent precursors of 24 nt siRNAs guiding de novo DNA methylation in Arabidopsis. eLife 4:e09591
Bouhouche K, Gout JF, Kapusta A, Betermier M, Meyer E (2011) Functional specialization of Piwi proteins in Paramecium tetraurelia from post-transcriptional gene silencing to genome remodelling. Nucleic Acids Res 39(10):4249–4264. https://doi.org/10.1093/nar/gkq1283
Bracht JR, Perlman DH, Landweber LF (2012) Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol 13(10):R99. https://doi.org/10.1186/gb-2012-13-10-r99
Calarco JP, Borges F, Donoghue MT, Van Ex F, Jullien PE, Lopes T, Gardner R, Berger F, Feijo JA, Becker JD et al (2012) Reprogramming of DNA methylation in pollen guides epigenetic inheritance via small RNA. Cell 151(1):194–205. https://doi.org/10.1016/j.cell.2012.09.001
Cao X, Aufsatz W, Zilberman D, Mette MF, Huang MS, Matzke M, Jacobsen SE (2003) Role of the DRM and CMT3 methyltransferases in RNA-directed DNA methylation. Curr Biol 13(24):2212–2217. https://doi.org/10.1016/j.cub.2003.11.052
Chalker DL, Fuller P, Yao MC (2005) Communication between parental and developing genomes during tetrahymena nuclear differentiation is likely mediated by homologous RNAs. Genetics 169(1):149–160. https://doi.org/10.1534/genetics.104.032300
Chalker DL, Yao MC (2001) Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev 15(10):1287–1298. https://doi.org/10.1101/gad.884601
Chen X, Bracht JR, Goldman AD, Dolzhenko E, Clay DM, Swart EC, Perlman DH, Doak TG, Stuart A, Amemiya CT, Sebra RP, Landweber LF (2014) The architecture of a scrambled genome reveals massive levels of genomic rearrangement during development. Cell 158(5):1187–1198. https://doi.org/10.1016/j.cell.2014.07.034
Cheng CY, Vogt A, Mochizuki K, Yao MC (2010) A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell 21(10):1753–1762. https://doi.org/10.1091/mbc.E09-12-1079
Du J, Johnson LM, Jacobsen SE, Patel DJ (2015) DNA methylation pathways and their crosstalk with histone methylation. Nat Rev Mol Cell Biol 16(9):519–532. https://doi.org/10.1038/nrm4043
Dubois E, Bischerour J, Marmignon A, Mathy N, Regnier V, Betermier M (2012) Transposon invasion of the Paramecium germline genome countered by a domesticated PiggyBac transposase and the NHEJ pathway. Int J Evol Biol 2012:436196
Duharcourt S, Butler A, Meyer E (1995) Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev 9(16):2065–2077. https://doi.org/10.1101/gad.9.16.2065
Duret L, Cohen J, Jubin C, Dessen P, Gout JF, Mousset S, Aury JM, Jaillon O, Noel B, Arnaiz O, Betermier M, Wincker P, Meyer E, Sperling L (2008) Analysis of sequence variability in the macronuclear DNA of Paramecium tetraurelia: a somatic view of the germline. Genome Res 18(4):585–596. https://doi.org/10.1101/gr.074534.107
Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, Tallon LJ, Delcher AL, Salzberg SL, Silva JC, Haas BJ, Majoros WH, Farzad M, Carlton JM, Smith RK, Garg J, Pearlman RE, Karrer KM, Sun L, Manning G, Elde NC, Turkewitz AP, Asai DJ, Wilkes DE, Wang Y, Cai H, Collins K, Stewart BA, Lee SR, Wilamowska K, Weinberg Z, Ruzzo WL, Wloga D, Gaertig J, Frankel J, Tsao CC, Gorovsky MA, Keeling PJ, Waller RF, Patron NJ, Cherry JM, Stover NA, Krieger CJ, del Toro C, Ryder HF, Williamson SC, Barbeau RA, Hamilton EP, Orias E (2006) Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol 4(9):e286. https://doi.org/10.1371/journal.pbio.0040286
Fang W, Wang X, Bracht JR, Nowacki M, Landweber LF (2012) Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell 151(6):1243–1255. https://doi.org/10.1016/j.cell.2012.10.045
Fass JN, Joshi NA, Couvillion MT, Bowen J, Gorovsky MA, Hamilton EP, Orias E, Hong K, Coyne RS, Eisen JA, Chalker DL, Lin D, Collins K (2011) Genome-scale analysis of programmed DNA elimination sites in Tetrahymena thermophila. G3 (Bethesda) 1(6):515–522. https://doi.org/10.1534/g3.111.000927
Furrer DI, Swart EC, Kraft MF, Sandoval PY, Nowacki M (2017) Two sets of Piwi proteins are involved in distinct sRNA pathways leading to elimination of germline-specific DNA. Cell Rep 20(2):505–520. https://doi.org/10.1016/j.celrep.2017.06.050
Guerin F, Arnaiz O, Boggetto N, Denby Wilkes C, Meyer E, Sperling L, Duharcourt S (2017) Flow cytometry sorting of nuclei enables the first global characterization of Paramecium germline DNA and transposable elements. BMC Genomics 18(1):327. https://doi.org/10.1186/s12864-017-3713-7
Haag JR, Pikaard CS (2011) Multisubunit RNA polymerases IV and V: purveyors of non-coding RNA for plant gene silencing. Nat Rev Mol Cell Biol 12(8):483–492. https://doi.org/10.1038/nrm3152
Haag JR, Pontes O, Pikaard CS (2009) Metal A and metal B sites of nuclear RNA polymerases Pol IV and Pol V are required for siRNA-dependent DNA methylation and gene silencing. PLoS One 4(1):e4110. https://doi.org/10.1371/journal.pone.0004110
Hamilton, E.P., Kapusta, A., Huvos, P.E., Bidwell, S.L., Zafar, N., Tang, H., Hadjithomas, M., Krishnakumar, V., Badger, J.H., Caler, E.V., et al. (2016). Structure of the germline genome of Tetrahymena thermophila and relationship to the massively rearranged somatic genome. Elife 5
Havecker ER, Wallbridge LM, Hardcastle TJ, Bush MS, Kelly KA, Dunn RM, Schwach F, Doonan JH, Baulcombe DC (2010) The Arabidopsis RNA-directed DNA methylation argonautes functionally diverge based on their expression and interaction with target loci. Plant Cell 22(2):321–334. https://doi.org/10.1105/tpc.109.072199
Heard E, Martienssen RA (2014) Transgenerational epigenetic inheritance: myths and mechanisms. Cell 157(1):95–109. https://doi.org/10.1016/j.cell.2014.02.045
Herr AJ, Jensen MB, Dalmay T, Baulcombe DC (2005) RNA polymerase IV directs silencing of endogenous DNA. Science 308(5718):118–120. https://doi.org/10.1126/science.1106910
Hsieh TF, Ibarra CA, Silva P, Zemach A, Eshed-Williams L, Fischer RL, Zilberman D (2009) Genome-wide demethylation of Arabidopsis endosperm. Science 324(5933):1451–1454. https://doi.org/10.1126/science.1172417
Johnson LM, Bostick M, Zhang X, Kraft E, Henderson I, Callis J, Jacobsen SE (2007) The SRA methyl-cytosine-binding domain links DNA and histone methylation. Curr Biol 17(4):379–384. https://doi.org/10.1016/j.cub.2007.01.009
Jullien PE, Susaki D, Yelagandula R, Higashiyama T, Berger F (2012) DNA methylation dynamics during sexual reproduction in Arabidopsis thaliana. Curr Biol 22(19):1825–1830. https://doi.org/10.1016/j.cub.2012.07.061
Kanno T, Bucher E, Daxinger L, Huettel B, Kreil DP, Breinig F, Lind M, Schmitt MJ, Simon SA, Gurazada SG et al (2010) RNA-directed DNA methylation and plant development require an IWR1-type transcription factor. EMBO Rep 11(1):65–71. https://doi.org/10.1038/embor.2009.246
Kanno T, Huettel B, Mette MF, Aufsatz W, Jaligot E, Daxinger L, Kreil DP, Matzke M, Matzke AJ (2005) Atypical RNA polymerase subunits required for RNA-directed DNA methylation. Nat Genet 37(7):761–765. https://doi.org/10.1038/ng1580
Kapusta A, Matsuda A, Marmignon A, Ku M, Silve A, Meyer E, Forney JD, Malinsky S, Betermier M (2011) Highly precise and developmentally programmed genome assembly in Paramecium requires ligase IV-dependent end joining. PLoS Genet 7(4):e1002049. https://doi.org/10.1371/journal.pgen.1002049
Kasschau KD, Fahlgren N, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Carrington JC (2007) Genome-wide profiling and analysis of Arabidopsis siRNAs. PLoS Biol 5(3):e57. https://doi.org/10.1371/journal.pbio.0050057
Kataoka K, Mochizuki K (2011) Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism. Adv Exp Med Biol 722:156–173. https://doi.org/10.1007/978-1-4614-0332-6_10
Khurana JS, Wang X, Chen X, Perlman DH, Landweber LF (2014) Transcription-independent functions of an RNA polymerase II subunit, Rpb2, during genome rearrangement in the ciliate, Oxytricha trifallax. Genetics 197(3):839–849. https://doi.org/10.1534/genetics.114.163279
Kornberg RD (2007) The molecular basis of eukaryotic transcription. Proc Natl Acad Sci U S A 104(32):12955–12961. https://doi.org/10.1073/pnas.0704138104
Law JA, Jacobsen SE (2010) Establishing, maintaining and modifying DNA methylation patterns in plants and animals. Nat Rev Genet 11(3):204–220. https://doi.org/10.1038/nrg2719
Lepere G, Betermier M, Meyer E, Duharcourt S (2008) Maternal noncoding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev 22(11):1501–1512. https://doi.org/10.1101/gad.473008
Lhuillier-Akakpo M, Frapporti A, Denby Wilkes C, Matelot M, Vervoort M, Sperling L, Duharcourt S (2014) Local effect of enhancer of zeste-like reveals cooperation of epigenetic and cis-acting determinants for zygotic genome rearrangements. PLoS Genet 10(9):e1004665. https://doi.org/10.1371/journal.pgen.1004665
Li CF, Pontes O, El-Shami M, Henderson IR, Bernatavichute YV, Chan SW, Lagrange T, Pikaard CS, Jacobsen SE (2006) An ARGONAUTE4-containing nuclear processing center colocalized with Cajal bodies in Arabidopsis thaliana. Cell 126(1):93–106. https://doi.org/10.1016/j.cell.2006.05.032
Li J, Yang Z, Yu B, Liu J, Chen X (2005) Methylation protects miRNAs and siRNAs from a 3′-end uridylation activity in Arabidopsis. Curr Biol 15(16):1501–1507. https://doi.org/10.1016/j.cub.2005.07.029
Lindblad, K.A., Bracht, J.R., Williams, A.E., and Landweber, L. (2017). Thousands of RNA-cached copies of whole chromosomes are present in the ciliate Oxytricha during development. RNA
Lindroth AM, Cao X, Jackson JP, Zilberman D, McCallum CM, Henikoff S, Jacobsen SE (2001) Requirement of CHROMOMETHYLASE3 for maintenance of CpXpG methylation. Science 292(5524):2077–2080. https://doi.org/10.1126/science.1059745
Lindroth AM, Shultis D, Jasencakova Z, Fuchs J, Johnson L, Schubert D, Patnaik D, Pradhan S, Goodrich J, Schubert I, Jenuwein T, Khorasanizadeh S, Jacobsen SE (2004) Dual histone H3 methylation marks at lysines 9 and 27 required for interaction with CHROMOMETHYLASE3. EMBO J 23(21):4286–4296. https://doi.org/10.1038/sj.emboj.7600430
Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD (2007) RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev 21(12):1530–1545. https://doi.org/10.1101/gad.1544207
Lu J, Zhang C, Baulcombe DC, Chen ZJ (2012) Maternal siRNAs as regulators of parental genome imbalance and gene expression in endosperm of Arabidopsis seeds. Proc Natl Acad Sci U S A 109(14):5529–5534. https://doi.org/10.1073/pnas.1203094109
Malone CD, Anderson AM, Motl JA, Rexer CH, Chalker DL (2005) Germ line transcripts are processed by a Dicer-like protein that is essential for developmentally programmed genome rearrangements of Tetrahymena thermophila. Mol Cell Biol 25(20):9151–9164. https://doi.org/10.1128/MCB.25.20.9151-9164.2005
Matzke MA, Mosher RA (2014) RNA-directed DNA methylation: an epigenetic pathway of increasing complexity. Nat Rev Genet 15(6):394–408. https://doi.org/10.1038/nrg3683
Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA (2002) Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in tetrahymena. Cell 110(6):689–699. https://doi.org/10.1016/S0092-8674(02)00909-1
Mochizuki K, Gorovsky MA (2004a) Conjugation-specific small RNAs in Tetrahymena have predicted properties of scan (scn) RNAs involved in genome rearrangement. Genes Dev 18(17):2068–2073. https://doi.org/10.1101/gad.1219904
Mochizuki K, Gorovsky MA (2004b) RNA polymerase II localizes in Tetrahymena thermophila meiotic micronuclei when micronuclear transcription associated with genome rearrangement occurs. Eukaryot Cell 3(5):1233–1240. https://doi.org/10.1128/EC.3.5.1233-1240.2004
Mochizuki K, Gorovsky MA (2005) A Dicer-like protein in Tetrahymena has distinct functions in genome rearrangement, chromosome segregation, and meiotic prophase. Genes Dev 19(1):77–89. https://doi.org/10.1101/gad.1265105
Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC (2009) Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature 460(7252):283–286. https://doi.org/10.1038/nature08084
Neeb ZT, Hogan DJ, Katzman S, Zahler AM (2017) Preferential expression of scores of functionally and evolutionarily diverse DNA and RNA-binding proteins during Oxytricha trifallax macronuclear development. PLoS One 12(2):e0170870. https://doi.org/10.1371/journal.pone.0170870
Noto T, Kataoka K, Suhren JH, Hayashi A, Woolcock KJ, Gorovsky MA, Mochizuki K (2015) Small-RNA-mediated genome-wide trans-recognition network in Tetrahymena DNA elimination. Mol Cell 59(2):229–242. https://doi.org/10.1016/j.molcel.2015.05.024
Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF (2008) RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature 451(7175):153–158. https://doi.org/10.1038/nature06452
Onodera Y, Haag JR, Ream T, Costa Nunes P, Pontes O, Pikaard CS (2005) Plant nuclear RNA polymerase IV mediates siRNA and DNA methylation-dependent heterochromatin formation. Cell 120(5):613–622. https://doi.org/10.1016/j.cell.2005.02.007
Pontier D, Yahubyan G, Vega D, Bulski A, Saez-Vasquez J, Hakimi MA, Lerbs-Mache S, Colot V, Lagrange T (2005) Reinforcement of silencing at transposons and highly repeated sequences requires the concerted action of two distinct RNA polymerases IV in Arabidopsis. Genes Dev 19(17):2030–2040. https://doi.org/10.1101/gad.348405
Prescott DM (1994) The DNA of ciliated protozoa. Microbiol Rev 58(2):233–267
Qi Y, Denli AM, Hannon GJ (2005) Biochemical specialization within Arabidopsis RNA silencing pathways. Mol Cell 19(3):421–428. https://doi.org/10.1016/j.molcel.2005.06.014
Qi Y, He X, Wang XJ, Kohany O, Jurka J, Hannon GJ (2006) Distinct catalytic and non-catalytic roles of ARGONAUTE4 in RNA-directed DNA methylation. Nature 443(7114):1008–1012. https://doi.org/10.1038/nature05198
Quadrana L, Colot V (2016) Plant transgenerational epigenetics. Annu Rev Genet 50(1):467–491. https://doi.org/10.1146/annurev-genet-120215-035254
Sandoval PY, Swart EC, Arambasic M, Nowacki M (2014) Functional diversification of Dicer-like proteins and small RNAs required for genome sculpting. Dev Cell 28(2):174–188. https://doi.org/10.1016/j.devcel.2013.12.010
Stroud H, Do T, Du J, Zhong X, Feng S, Johnson L, Patel DJ, Jacobsen SE (2014) Non-CG methylation patterns shape the epigenetic landscape in Arabidopsis. Nat Struct Mol Biol 21(1):64–72. https://doi.org/10.1038/nsmb.2735
Stroud H, Greenberg MV, Feng S, Bernatavichute YV, Jacobsen SE (2013) Comprehensive analysis of silencing mutants reveals complex regulation of the Arabidopsis methylome. Cell 152(1-2):352–364. https://doi.org/10.1016/j.cell.2012.10.054
Swart EC, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, Jung S, Fulton RS, Ly A, McGrath S, Haub K, Wiggins JL, Storton D, Matese JC, Parsons L, Chang WJ, Bowen MS, Stover NA, Jones TA, Eddy SR, Herrick GA, Doak TG, Wilson RK, Mardis ER, Landweber LF (2013) The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol 11(1):e1001473. https://doi.org/10.1371/journal.pbio.1001473
Taverna SD, Coyne RS, Allis CD (2002) Methylation of histone h3 at lysine 9 targets programmed DNA elimination in tetrahymena. Cell 110(6):701–711. https://doi.org/10.1016/S0092-8674(02)00941-8
Tucker SL, Reece J, Ream TS, Pikaard CS (2010) Evolutionary history of plant multisubunit RNA polymerases IV and V: subunit origins via genome-wide and segmental gene duplications, retrotransposition, and lineage-specific subfunctionalization. Cold Spring Harb Symp Quant Biol 75(0):285–297. https://doi.org/10.1101/sqb.2010.75.037
Vongs A, Kakutani T, Martienssen RA, Richards EJ (1993) Arabidopsis thaliana DNA methylation mutants. Science 260(5116):1926–1928. https://doi.org/10.1126/science.8316832
Wierzbicki AT, Haag JR, Pikaard CS (2008) Noncoding transcription by RNA polymerase Pol IVb/Pol V mediates transcriptional silencing of overlapping and adjacent genes. Cell 135(4):635–648. https://doi.org/10.1016/j.cell.2008.09.035
Wierzbicki AT, Ream TS, Haag JR, Pikaard CS (2009) RNA polymerase V transcription guides ARGONAUTE4 to chromatin. Nat Genet 41(5):630–634. https://doi.org/10.1038/ng.365
Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC (2004) Genetic and functional diversification of small RNA pathways in plants. PLoS Biol 2(5):E104. https://doi.org/10.1371/journal.pbio.0020104
Yao MC, Chao JL (2005) RNA-guided DNA deletion in Tetrahymena: an RNAi-based mechanism for programmed genome rearrangements. Annu Rev Genet 39(1):537–559. https://doi.org/10.1146/annurev.genet.39.073003.095906
Yu B, Yang Z, Li J, Minakhina S, Yang M, Padgett RW, Steward R, Chen X (2005) Methylation as a crucial step in plant microRNA biogenesis. Science 307(5711):932–935. https://doi.org/10.1126/science.1107130
Zahler AM, Neeb ZT, Lin A, Katzman S (2012) Mating of the stichotrichous ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS One 7(8):e42371. https://doi.org/10.1371/journal.pone.0042371
Zhai J, Bischof S, Wang H, Feng S, Lee TF, Teng C, Chen X, Park SY, Liu L, Gallego-Bartolome J et al (2015) A one precursor one siRNA model for Pol IV-dependent siRNA biogenesis. Cell 163(2):445–455. https://doi.org/10.1016/j.cell.2015.09.032
Zhang X, Henderson IR, Lu C, Green PJ, Jacobsen SE (2007) Role of RNA polymerase IV in plant small RNA metabolism. Proc Natl Acad Sci U S A 104(11):4536–4541. https://doi.org/10.1073/pnas.0611456104
Zhong X, Du J, Hale CJ, Gallego-Bartolome J, Feng S, Vashisht AA, Chory J, Wohlschlegel JA, Patel DJ, Jacobsen SE (2014) Molecular mechanism of action of plant DRM de novo DNA methyltransferases. Cell 157(5):1050–1060. https://doi.org/10.1016/j.cell.2014.03.056
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
This research does not include human participants or animals.
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Neeb, Z.T., Nowacki, M. RNA-mediated transgenerational inheritance in ciliates and plants. Chromosoma 127, 19–27 (2018). https://doi.org/10.1007/s00412-017-0655-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-017-0655-4