Abstract
The zebra finch (Taeniopygia guttata) germline-restricted chromosome (GRC) is the largest chromosome and has a unique system of transmission in germ cells. In the male, the GRC exists as a single heterochromatic chromosome in the germline and is eliminated from nuclei in late spermatogenesis. In the female, the GRC is bivalent and euchromatic and experiences recombination. These characteristics suggest a female-specific or female-beneficial function of the GRC. To shed light on the function of GRC, we cloned a portion of the GRC using random amplified polymorphic DNA–polymerase chain reaction and analyzed it using molecular genetic and cytogenetic methods. The GRC clone hybridized strongly to testis but not blood DNA in genomic Southern blots. In fluorescent in situ hybridization analysis on meiotic chromosomes from synaptonemal complex spreads, the probe showed hybridization across a large area of the GRC, suggesting that it contains repetitive sequences. We isolated a sequence homologous to the GRC from zebra finch chromosome 3 and a region of chicken chromosome 1 that is homologous to zebra finch chromosome 3; the phylogenetic analysis of these three sequences suggested that the GRC sequence and the zebra finch chromosome 3 sequence are most closely related. Thus, the GRC sequences likely originated from autosomal DNA and have evolved after the galliform–passeriform split. The present study provides a foundation for further study of the intriguing GRC.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
The number of chromosomes differs between somatic and germline cells in several organisms; Acricotopus (Staiber 1987), sciarid flies (Goday and Esteban 2001), and some hagfish species (Kohno et al. 1986; Kubota et al. 1993). The extra chromosomes in the germline, germline-restricted chromosomes (GRCs), are eliminated during early embryogenesis and do not exist in somatic cells. Thus, unlike A chromosomes, which are regular elements following Mendelian laws, GRCs have unique characteristics. In Acricotopus lucidus, there are nine different types of GRC distinguished by G-banding, which exist as various combinations in individuals; some are missing and others are present as multiple copies (Staiber 1988). Fluorescent in situ hybridization (FISH) experiments confirm that these nine GRCs have homology to A chromosome sequences and sometimes have pairing-like associations to A chromosomes (Staiber and Schiffkowski 2000; Staiber and Wahl 2002). The homology of a GRC and its homologous A chromosome sequences are high, which suggests an A chromosomal origin of GRC sequences (Staiber 2004). The sequences specific to GRCs have been tandemly repeated and are mainly located on the centromeric heterochromatin regions (Staiber et al. 1997; Staiber 2002). In most cases, the GRC of A. lucidus is eliminated in one mitosis and move to the pole cell, but some populations escape this first division and are eliminated in the following mitoses (Staiber 2006). In hagfish, the GRC DNA sequences cloned from Eptatretus okinoseanus, Paramyxine atami, Eptatretus cirrhatus, and Eptatretus burgeri also showed tandem repeats, and interestingly, some of the repeat sequences are conserved across species, suggesting their functional importance (Kubota et al. 1993, 1997, 2001; Goto et al. 1998; Nabeyama et al. 2000).
The zebra finch has 80 chromosomes including seven pairs of macrochromosomes, heteromorphic ZW sex chromosomes, and 32 pairs of microchromosomes (Pigozzi and Solari 1998; Itoh and Arnold 2005). This oscine Passerine songbird is the second bird species for which the whole genome has been sequenced (http://genome.ucsc.edu) and is important for studies of behavioral neurobiology, sexual differentiation, and comparative genomics (Marler 1990; Nottebohm 2002; Konishi 1994; Arnold 2002; Itoh et al. 2007, Replogle et al. 2008). The avian chromosomes are generally highly conserved in gene content, so that there has been relatively little rearrangement of the genome in the last 100Myr since the divergence of the more ancestral ratites (Struthioniformes, flightless birds) and the rest of the birds, the carinates (Shetty et al. 1999). Based on cytogenetic analysis, the zebra finch and chicken (Gallus gallus) show conservation of gene content of all of the macrochromosomes although there are several rearrangements (Pigozzi and Solari 1998; Itoh and Arnold 2005). Interestingly, Pigozzi and Solari (1998, 2005) reported a GRC in the zebra finch. This chromosome is the largest chromosome in zebra finch. One species related to zebra finch also carries the GRC (Pigozzi, unpublished results). The transmission of this chromosome is not Mendelian and its meiotic behavior differs in male and female germ cells. In testis, the GRC exists as a single heterochromatic chromosome that is eliminated from nuclei during meiosis. In contrast, in the female, the GRC is a euchromatic bivalent that recombines asexually during meiosis. Since the GRC is excluded from mature male germ cells and does not exist in sperm, there must be duplication of the GRC chromosome originating from the female at some point after fertilization so that recombination occurs in female germ cells. Thus, the maintenance/transmission of GRC is a side event in females. These properties suggest that the GRC is functionally important in female germ cells, for example, in female gametogenesis. Or, it could contain copies of genes homologous to Z-linked genes that would otherwise show hemizygous insufficiency in females because of a lack of effective Z chromosome dosage compensation in birds (Itoh et al. 2007; Ellegren et al. 2007; Arnold et al. 2008). Thus, the GRC might be seen as an adaptation to compensate in a tissue-specific manner for sex differences in Z gene dosage.
In this study, we cloned part of the zebra finch GRC sequence from testis DNA and characterized it by molecular and cytogenetic methods. The GRC sequence we cloned was distributed across a large region of the GRC, suggesting that the GRC contains repetitive elements. We also found sequence homologous to the GRC on the short arm of zebra finch chromosome 3, which was strongly conserved on chicken chromosome 1, suggesting the A chromosome origin of this GRC sequence.
Materials and methods
Preparation of genomic DNA
High-molecular-weight genomic DNA was isolated from nuclei of erythrocytes or several tissue samples. Briefly, the nuclei of blood cells or 1-mm3-sized tissue samples were suspended in 10 mM Tris–HCl (pH 8.0) and 100 mM ethylenediaminetetraacetic acid (EDTA; pH 8.0) and incubated in the presence of 0.5% sodium dodecyl sulfate (SDS) and 100 µg/mL proteinase K at 50°C overnight. The mixture was extracted successively with phenol saturated with TE [10 mM Tris–HCl (pH 8.0), 1 mM EDTA] and chloroform. DNA was ethanol precipitated from the supernatant, dissolved in TE, and stored at 4°C.
Random amplified polymorphic DNA PCR and cloning
After testing 52 arbitrary polymerase chain reaction (PCR) primers, primer number 5 (5′-GAGCAGGCTG-3′) was found to amplify testis-specific sequences (TRAP5) from zebra finch genomic DNA. The PCR was carried out in a 25-µL mixture containing 0.2 mM each of dNTPs, 3 mM MgCl2, 0.4 µM each of primers, 10 ng genomic DNA, 0.25 µL of Taq polymerase (Invitrogen), and one tenth volume of 10x Taq buffer (Invitrogen). The PCR reaction was held at 94°C for 2 min before the cycling reaction of two cycles of 94°C for 2 min/36°C for 2 min/72°C for 2 min, 38 cycles of 94°C for 30 s/36°C for 30 s/72°C for 30 s followed by a single 7 min period at 72°C. Ten microliters of the PCR product was separated by 1.5 % agarose gel electrophoresis in 1x TAE (10 mM Tris, 4 mM acetic acid, 0.5 mM EDTA) at 80 V. The PCR products were cloned into pGEM-T Easy (Promega) and sequenced.
PCR amplification of GRC sequence using specific primers
The PCR was carried out in a 25-µL mixture containing 0.2 mM each of dNTPs, 2 mM MgCl2, 0.4 µM each of primers, 10 ng genomic DNA, 0.25 µL of Taq polymerase (Invitrogen), and one tenth volume of 10x Taq buffer (Invitrogen). The sequences of primers were 27L4-F2 (5′-ATCTGAGGGTCCAAGCAATG-3′) and 27L4-R2 (5′-ATTGTCACGGGAAAAGTCCA-3′). The PCR reaction started at 95°C for 4 min before the cycling reaction of 40 cycles of 95°C for 45 s/64°C for 30 s/72°C for 1 min and then followed by single reaction at 72°C for 7 min. Ten microliters of the PCR reaction mixture was separated by 1.5% agarose gel electrophoresis in 1x TAE at 80 V.
PCR amplification of TRAP5 homologous sequence on A chromosome
The PCR was carried out in a 25-µL mixture containing 0.2 mM each of dNTPs, 2 mM MgCl2, 0.4 µM each of primers, 10 ng genomic DNA, 0.25 µL of Taq polymerase (Invitrogen), and one tenth volume of 10x Taq buffer (Invitrogen). The sequences of primers were TRAP5 F3 (5′-GAAGAAGGAAATCAGACTGGA-3′) and TRAP5RA1 (5′-TTCAGCTAAAAGCAGGAGTG-3′). The PCR reaction started at 95°C for 4 min before the cycling reaction of 40 cycles of 95°C for 45 s/59°C for 30 s/72°C for 1.5 min and then followed by a single cycle reaction at 72°C for 7 min. Ten microliters of the PCR reaction mixture was separated by 1.5% agarose gel electrophoresis in 1x TAE at 80 V.
Southern blot hybridization
Blood- or tissue-derived genomic DNA was digested with a restriction enzyme as indicated, electrophoresed in a 0.8% agarose gel (20 µg/lane), and transferred to Hybond-N+ membrane (Amersham Pharmacia Biotech) in 0.4 N NaOH and 0.6 mol/L NaCl. Hybridization with a 32P-labeled probe was carried out in a hybridization buffer [0.5 mol/L Na–phosphate buffer (pH 7.2), 7% SDS, 1 mmol/L EDTA] at 65°C. The membrane was washed in 2x SSC, 0.1% SDS at room temperature for 1 min twice and 65°C for 30 min twice, and subjected to autoradiography.
Lambda and BAC clones isolation and fluorescent in situ hybridization
27L4, 1328, and 13811 lambda DNA clones were isolated from a zebra finch testis genomic library using a TRAP5 fragment as a probe. For A chromosome homologs of TRAP5, zebra finch BAC clones (149E22, 027F04, and 081I17) were isolated from a zebra finch BAC library made by the Arizona Genomics Institute (http://www.genome.arizona.edu; Luo et al. 2006). BAC and lambda clones were used in FISH to probe mitotic chromosomes (Itoh and Arnold 2005) and male and female meiotic chromosomes along with immunostained synaptonemal complexes (SCs; Pigozzi 2007, 2008).
Results
Molecular cloning of zebra finch testis-specific sequence
To isolate the GRC-derived sequence, we performed random amplified polymorphic DNA–polymerase chain reaction (RAPD-PCR) on blood and testis genomic DNA isolated from the same animal to minimize polymorphic amplification. We chose testis DNA as the source for GRC sequences because the germline cell population relative to mitotic cells is significantly higher than in the ovary. Out of 52 arbitrary PCR primers used, primer number 5 amplified 0.9 kb PCR product (TRAP5) from testis genomic DNA sample but not blood in samples from two different animals (Fig. 1a). We further tested PCR amplification using primer number 5 in blood, brain, liver, and gonad genomic DNA from different male and female samples. Testis-specific amplification was confirmed, but no amplification was found in the ovary (Fig. 1b). Using GRC-specific primers, GRC DNA was amplified only from the testis, not the ovary (Fig. 2). DNA quality of samples was confirmed by amplification of spindlin DNA (data not shown; Itoh et al. 2001). The lack of amplification of GRC sequences from ovary DNA has at least three possible explanations: (1) GRC DNA in the testis and ovary are different. (2) The sequence variation among GRCs is large enough that the primer sequences did not match in the females used. (3) The proportion of germline cells in the ovary is small enough that the level of GRC DNA in the sample was insufficient for amplification of TRAP5. Because the probe derived from TRAP5 hybridized to female meiotic chromosome (see below), we favor the last explanation.
Testis specificity of the TRAP5 sequence was further confirmed by Southern blot hybridization to male/female blood DNA and testis DNA (Fig. 3). The GRC bands recognized in an equally loaded genomic Southern blot (Fig. 3a) were particularly heavy in testis samples (~2 kb in HindIII-digested and ~3 kb in HaeIII-digested DNA; Fig. 3b). The strength of these signals suggested that the TRAP5 probe sequence existed as repeats in the testis samples. The blood samples had much weaker signals that were comparable in size to those from the testis, suggesting the existence of the homologous sequences on A chromosome(s). These weaker signals in blood samples were more obvious when we exposed the autoradiograph for a longer period (Fig. 3c).
TRAP5 sequence and its homologous sequence on A chromosome
The sequence of TRAP5 (Fig. 4) did not show any strong homology to any genes or sequences in the DDBJ/EMBL/GenBank database. Based on the idea that GRC homologous sequences exist on A chromosomes (Fig. 3), we screened a zebra finch liver DNA BAC library, which should not contain GRC, with the TRAP5 probe and isolated a putative A chromosome-related GRC homologous BAC DNA clone (147E22). FISH mapping of 147E22 BAC clone to zebra finch metaphase chromosome sets showed the signal localization on the short arm of chromosome 3 (Fig. 5). We also cloned this chromosome 3 sequence by PCR from blood DNA using the PCR primers for GRC sequence (TRAP5 F3 and TRAP5RA1). The amplified 1,470-bp fragment (TRAP5AH3) was highly homologous to the TRAP5 sequence (Fig. 6).
To determine if the TRAP5 sequence represents a complete tandem repeat unit, we isolated a larger genomic fragment of the GRC by screening of zebra finch testis lambda genomic library (27L4, GenBank accession number FJ609199). Genome walking further identified two other lambda DNA clones on either side of 27L4 (1328 and 13811; Fig. 7). The 27L4 DNA sequence aligned well to the chicken genome in the region of 97,606,000–97,629,439 bp of chromosome 1, as well as to a zebra finch chromosome 3 genome sequence (Fig. 8a). Of the two autosomal sequences, the zebra finch chromosome 3 sequence showed significantly higher homology to GRC than did the chicken sequence. Two short sequences located on 8,754–8,826 and 14,781–14,885 bp of 27L4 (GenBank accession number FJ609199; Fig. 7) are sparse repetitive sequences which showed homology to sequences on chicken chromosome 1, 4, 5, 10, 12, 14, 15, and 16 (Fig. 8b). The calculation of evolutionary distance among chicken chromosome 1, zebra finch chromosome 3, and GRC sequences confirmed the closer relationship of GRC sequence to zebra finch chromosome 3 than to chicken chromosome 1 (Fig. 8c, d).
Physical map of GRC lambda DNA clone 27L4 and original RAPD product TRAP5. Two lambda DNA clones, 1328 and 13811, are found on the ends of 27L4. 27L4, 1328, and 13811 cover almost 45 kb in total. There are two predicted short-length sparse repetitive sequences on 8,754–8,826 and 14,781–14,885 bp. EXT32 is the exon trapping product
a Blast result of 18.8 kb TRAP5 lambda DNA sequence (27L4) against chicken chromosome 1 (97,606,077–97,626,571 bp; Ensemble chicken genome database) and zebra finch chromosome 3 genome sequence (http://genome.ucsc.edu/cgi-bin/hgGateway?db=taeGut1). The largest region of chicken Chr1 that does not align with TRAP5 is an unsequenced region of unknown size (Chr1 97,624,921–97,625,503 bp). b 27L4 localizes on chromosome 1 in chicken genome (box). Arrows show the localization of the homologous sequence of part of 27L4 on other chicken autosomes. c The phylogenetic tree constructed by maximum likelihood (http://atgc.lirmm.fr/phyml/). The parsimony estimation (http://mobyle.pasteur.fr/cgi-bin/MobylePortal/portal.py?form=dnapars) also showed the same result. Bar substitutions per nucleotide site
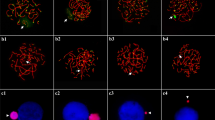
Meiotic chromosomal localization of 27L4 sequence
As previously reported, the GRC has a condensed state during male meiotic prophase and about half of metaphases II show an associated round condensed body, which presumably represents the heterochromatic form of the GRC (Pigozzi and Solari 1998, 2005). Figure 9a, b shows the heterochromatic GRC with DAPI staining in male meiotic nuclei. FISH mapping of the 27L4 sequence to the testis nuclei showed colocalization of the signal with the heterochromatic body in prophase spermatocytes (Fig. 9c, d). This GRC specificity was further confirmed by FISH mapping to male and female meiotic chromosomes (Figs. 9d and 10a, b). FISH mapping on female SC spreads shows that all three probes (27L4, 1328, and 13811) have repetitive sequence-like signals which are widely distributed over a large portion of GRC chromosome (Fig. 10a–e). The 27L4 probe also recognized an extra region near the centromere of the GRC which was not labeled by the other probes (Fig. 10c, e).
a Black–white reversed DAPI images of testis nuclei show highly condensed chromatin body. b, c FISH mapping of 27L4 lambda DNA to testis nuclei. The signals (red, c) were localized on the highly condensed chromatin body in testis nuclei. d FISH and immunostaining on a pachytene spermatocyte showing the extensive labeling (green) of the GRC (arrow) with the 13811 probe. The SCs and the single axis of the GRC are labeled with an antibody that recognizes the cohesin SMC3 (red). Scale bars = 10 µm
FISH mapping of 1328 (a), 13811 (b), and 27L4 (c) sequences (green) to female meiotic chromosomes represented by their SCs (red). a–c The green signal for all three probes locates on two major chromosome parts. d Partial view of a pachytene from oocyte showing the relative position of the probe (13811) signals with respect to the centromere (arrow). Centromeres were located using CREST serum and detected with a fluorescein isothiocyanate-labeled antibody. The arrowhead points at the ZW pair. e Schematic representation of the GRC regions (green) recognized by each of the three lambda DNA probes. The 27L4 probe recognizes an extra region to which the other probes do not hybridize. Bar = 10 µm
Discussion
We have isolated a zebra finch GRC-specific sequence from testis DNA using the RAPD-PCR technique and demonstrated its GRC specificity and repetitive nature. The existence of GRC homologous sequences on an A chromosome was suggested by Southern blot hybridization, and we cloned and mapped its homologous sequence on the short arm of zebra finch chromosome 3. These GRC and chromosome 3 sequences are highly conserved relative to each other and a homologous sequence was found on chicken chromosome 1 in a region homologous to zebra finch chromosome 3. The zebra finch GRC and chicken chromosome 1 sequence shared high homology and sequence order along a region of 18.8 kb. FISH mapping of the GRC sequence to meiotic chromosomes showed wide distribution of the sequence to a large portion of the GRC in both sexes, which suggested that this GRC chromosome contains a large amount of repeat sequence.
GRC sequences are thought to have evolved from A chromosome sequences by duplication, translocation/transposition, and accumulation of repetitive sequences. The zebra finch GRC is highly homologous to sequences on chromosome 3 which must be considered as ancestral to the GRC. This sequence is also conserved on chicken chromosome 1 although the chicken does not have a GRC (Pigozzi and Solari 1998), which raises the question whether zebra finch GRC has evolved as an independent event in zebra finch and/or related Passeriformes or whether it was distributed in numerous bird taxa but then subsequently lost in many lineages. More information concerning the distribution of the GRC is needed to decide this question. When we aligned the three sequences (GRC, zebra finch chromosome 3, and chicken chromosome 1) to each other by BLAST (http://blast.ncbi.nlm.nih.gov/bl2seq/wblast2.cgi), the GRC sequence had better alignment with zebra finch chromosome 3 than chicken chromosome 1 (Fig. 8a). Furthermore, the phylogenetic tree indicates a much closer evolutionary relationship between the GRC and zebra finch chromosome 3 (Fig. 8c, d). Thus, the zebra finch GRC sequence is much more closely related to its own chromosome 3 than are the two autosomal sequences in zebra finch and chicken, suggesting that the duplication of chromosome 3 sequences in the GRC in zebra finch lineage occurred after the split between chicken and zebra finch lineages. However, if the GRC mimics any function of chromosome 3, then their sequences could be kept similar by purifying selection.
Was duplication of chromosome 3 the starting point for GRC evolution? GRC evolution could be explained in two different mechanisms: (a) accidental whole or partial chromosome 3 duplication followed by specialization of the chromosome in a germline specific manner and (b) independent evolution of GRC followed by accidental partial duplication of chromosome 3 and translocation of that piece to the GRC. Most of the GRC sequence is not known. If much of the sequence is homologous to chromosome 3, hypothesis (a) would be supported. If not, then hypothesis (b) would be supported. In meiotic chromosome FISH mapping, the 20-kb 27L4 FISH probe hybridized to a large region of GRC, which suggests that this sequence encodes repetitive DNA. Since there is no tandem repeat unit observed in 27L4 and regions 3′ and 5′ to 27L4 showed a similar FISH pattern, we infer that either the repeat sequence are interspersed or any repeated elements show considerable rearrangement. This rearrangement is also suggestive in FISH signal pattern on SCs which showed discontinuous features in zebra finch, unlike the cloudy signal pattern in human and mouse which shows the probe hybridized to all the DNA loops around SCs (Froenicke et al. 2002; Codina-Pascual et al. 2006). Accumulation of repetitive sequences is a unique feature of GRCs, so it is possible that the ancestral zebra finch GRC sequence also followed that mechanism, probably because of the lack of recombination in spermatogenesis.
The GRC is univalent and heterochromatic in spermatogenetic germ cells, but in oocytes is bivalent and euchromatic and experiences recombination (Fig. 11). This pattern suggests that zebra finch GRC is functional in the female germline although we cannot exclude the possibility that some genes on GRC have functions in the male germline before the GRC is condensed and eliminated. To investigate if there is any functional gene expressed on GRC, we performed exon trapping (Itoh et al. 2001) on the 27L4 lambda clone and identified one predicted exon, which contained no open reading frame and for which we detected no expression (EXT32; Fig. 7). The Genscan gene identification software (http://bioweb.pasteur.fr/seqanal/interfaces/genscan-simple.html) also did not detect any meaningful open reading frame in the entire 27L4 sequence (18.8 kb). The 27L4 DNA clone used for exon trapping is highly repetitive on GRC, thus genes are not expected in that region. The existence of expressed sequences on other parts of the GRC is open for further investigation.
References
Arnold AP (2002) Concepts of genetic and hormonal induction of vertebrate sexual differentiation in the twentieth century, with special reference to the brain. In: Arnold AP, Etgen AM, Fahrbach SE, Rubin RT, Pfaff DW (eds) Hormones, brain, and behavior. Academic, San Diego, pp 105–135
Arnold AP, Itoh Y, Melamed E (2008) A bird's-eye view of sex chromosome dosage compensation. Annu Rev Genomics Hum Genet 9:109-127
Codina-Pascual M, Navarro J, Oliver-Bonet M, Kraus J, Speicher MR, Arango O, Egozcue J, Benet J (2006) Behaviour of human heterochromatic regions during the synapsis of homologous chromosomes. Mol Hum Reprod 12(2):123–133
Ellegren H, Hultin-Rosenberg L, Brunström B, Dencker L, Kultima K, Scholz B (2007) Faced with inequality: chicken do not have a general dosage compensation of sex-linked genes. BMC Biol 5:40
Froenicke L, Anderson LK, Wienberg J, Ashley T (2002) Male mouse recombination maps for each autosome identified by chromosome painting. Am J Hum Genet 71(6):1353–1368
Goday C, Esteban MR (2001) Chromosome elimination in sciarid flies. Bioessays 23(3):242–250
Goto Y, Kubota S, Kohno S (1998) Highly repetitive DNA sequences that are restricted to the germ line in the hagfish Eptatretus cirrhatus: a mosaic of eliminated elements. Chromosoma 107(1):17–32
Itoh Y, Hori T, Saitoh H, Mizuno S (2001) Chicken spindlin genes on W and Z chromosomes: transcriptional expression of both genes and dynamic behavior of spindlin in interphase and mitotic cells. Chromosome Res 9:283–299
Itoh Y, Arnold AP (2005) Chromosomal polymorphism and comparative painting analysis in the zebra finch. Chromosome Res 13:47–56
Itoh Y, Melamed E, Yang X, Kampf K, Wang S, Yehya N, Van Nas A, Replogle K, Band MR, Clayton DF, Schadt EE, Lusis AJ, Arnold AP (2007) Dosage compensation is less effective in birds than in mammals. J Biol 6(1):2
Kohno S, Nakai Y, Satoh S, Yoshida M, Kobayashi H (1986) Chromosome elimination in the Japanese hagfish, Eptatretus burgeri (Agnatha, Cyclostomata). Cytogenet Cell Genet 41(4): 209-214
Konishi M (1994) An outline of recent advances in birdsong neurobiology. Brain Behav Evol 44:279–285
Kubota S, Kuro-o M, Mizuno S, Kohno S (1993) Germ line-restricted, highly repeated DNA sequences and their chromosomal localization in a Japanese hagfish (Eptatretus okinoseanus). Chromosoma 102(3):163–173
Kubota S, Ishibashi T, Kohno S (1997) A germline restricted, highly repetitive DNA sequence in Paramyxine atami: an interspecifically conserved, but somatically eliminated, element. Mol Gen Genet 256(3):252–256
Kubota S, Takano J, Tsuneishi R, Kobayakawa S, Fujikawa N, Nabeyama M, Kohno S (2001) Highly repetitive DNA families restricted to germ cells in a Japanese hagfish (Eptatretus burgeri): a hierarchical and mosaic structure in eliminated chromosomes. Genetica 111(1–3):319–328
Luo M, Yu Y, Kim H, Kudrna D, Itoh Y, Agate RJ, Melamed E, Goicoechea JL, Talag J, Mueller C, Wang W, Currie J, Sisneros NB, Wing RA, Arnold AP (2006) Utilization of a zebra finch BAC library to determine the structure of an avian androgen receptor genomic region. Genomics 87(1):181–190
Marler P (1990) Song learning: the interface between behaviour and neuroethology. Philos Trans R Soc Lond B Biol Sci 329:109–114
Nabeyama M, Kubota S, Kohno S (2000) Concerted evolution of a highly repetitive DNA family in eptatretidae (Cyclostomata, agnatha) implies specifically differential homogenization and amplification events in their germ cells. J Mol Evol 50(2):154–69
Nottebohm F (2002) Neuronal replacement in adult brain. Brain Res Bull 57:737–749
Pigozzi MI, Solari AJ (1998) Germ cell restriction and regular transmission of an accessory chromosome that mimics a sex body in the zebra finch, Taeniopygia guttata. Chromosome Res 6:105–113
Pigozzi MI, Solari AJ (2005) The germ-line-restricted chromosome in the zebra finch: recombination in females and elimination in males. Chromosoma 114(6):403–409
Pigozzi MI (2007) Localization of single-copy sequences on chicken synaptonemal complex spreads using fluorescence in situ hybridization (FISH). Cytogenet Genome Res 119(1–2):105–112
Pigozzi MI (2008) Relationship between physical and genetic distances along the zebra finch Z chromosome. Chromosome Res 16:839–849
Replogle K, Arnold AP, Ball GF, Band M, Bensch S, Brenowitz EA, Dong S, Drnevich J, Ferris M, George JM, Gong G, Hasselquist D, Hernandez AG, Kim R, Lewin HA, Liu L, Lovell PV, Mello CV, Naurin S, Rodriguez-Zas S, Thimmapuram J, Wade J, Clayton DF (2008) The Songbird Neurogenomics (SoNG) Initiative: community-based tools and strategies for study of brain gene function and evolution. BMC Genomics 9:131
Shetty S, Griffin DK, Graves JA (1999) Comparative painting reveals strong chromosome homology over 80 million years of bird evolution. Chromosome Res 7:289–295
Staiber W (1987) Unusual germ line limited chromosomes in Acricotopus lucidus (Diptera, Chironomidae). Genome 29:702–705
Staiber W (1988) G-banding of germ line limited chromosomes in Acricotopus lucidus (Diptera, Chironomidae). Chromosoma 97:231–234
Staiber W, Wech I, Preiss A (1997) Isolation and chromosomal localization of a germ line-specific highly repetitive DNA family in Acricotopus lucidus (Diptera, Chironomidae). Chromosoma 106(5):267–275
Staiber W, Schiffkowski C (2000) Structural evolution of the germ line-limited chromosomes in Acricotopus. Chromosoma 109:343–349
Staiber W, Wahl S (2002) Painting analysis of meiotic metaphase I configurations of the germ line-limited chromosomes in Acricotopus. Chromosome Res 10(2):101–108
Staiber W (2002) Isolation of a new germ line-specific repetitive DNA family in Acricotopus by microdissection of polytenized germ line-limited chromosome sections from a permanent larval salivary gland preparation. Cytogenet Genome Res 98(2–3):210–215
Staiber W (2004) Molecular evolution of homologous gene sequences in germline-limited and somatic chromosomes of Acricotopus. Genome 47(4):732–741
Staiber W (2006) Chromosome elimination in germ line-soma differentiation of Acricotopus lucidus (Diptera, Chironomidae). Genome 49(3):269–274
Acknowledgements
This study was supported by NIH grant DC00217 to A.P. Arnold and a Yamada Science Foundation grant to Y. Itoh. The zebra finch chromosome 3 sequence was determined by the Washington University Genome Center under the supervision of Dr. Wes Warren. We thank Dr. Grace Xiao for the helpful suggestions.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by L. Comai.
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Itoh, Y., Kampf, K., Pigozzi, M.I. et al. Molecular cloning and characterization of the germline-restricted chromosome sequence in the zebra finch. Chromosoma 118, 527–536 (2009). https://doi.org/10.1007/s00412-009-0216-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-009-0216-6