Abstract
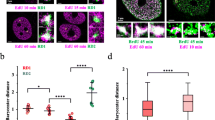
To study when and where active genes replicated in early S phase are transcribed, a series of pulse-chase experiments are performed to label replicating chromatin domains (RS) in early S phase and subsequently transcription sites (TS) after chase periods of 0 to 24 h. Surprisingly, transcription activity throughout these chase periods did not show significant colocalization with early RS chromatin domains. Application of novel image segmentation and proximity algorithms, however, revealed close proximity of TS with the labeled chromatin domains independent of chase time. In addition, RNA polymerase II was highly proximal and showed significant colocalization with both TS and the chromatin domains. Based on these findings, we propose that chromatin activated for transcription dynamically unfolds or “loops out” of early RS chromatin domains where it can interact with RNA polymerase II and other components of the transcriptional machinery. Our results further suggest that the early RS chromatin domains are transcribing genes throughout the cell cycle and that multiple chromatin domains are organized around the same transcription factory.








Similar content being viewed by others
References
Albiez H, Cremer M, Tiberi C, Vecchio L, Schermelleh L, Dittrich S, Kupper K, Joffe B, Thormeyer T, von Hase J, Yang S, Rohr K, Leonhardt H, Solovei I, Cremer C, Fakan S, Cremer T (2006) Chromatin domains and the interchromatin compartment form structurally defined and functionally interacting nuclear networks. Chromosome Res 14:707–733
Barber CB, Dobkin DP, Huhdanpaa HT (1996) The Quickhull algorithm for convex hulls. ACM Trans Math Softw 22:469–483
Berezney R (2002) Regulating the mammalian genome: the role of nuclear architecture. Adv Enzyme Regul 42:39–52
Berezney R, Wei X (1998) The new paradigm: integrating genomic function and nuclear architecture. J Cell Biochem 30–31:238–242
Berezney R, Mortillaro MJ, Ma H, Wei X, Samarabandu J (1995) The nuclear matrix: a structural milieu for genomic function. Int Rev Cyt 162A:1–65
Berezney R, Dubey DD, Huberman JA (2000) Heterogeneity of eukaryotic replicons, replicon clusters, and replication foci. Chromosoma 108:471–484
Berezney R, Malyavantham KS, Pliss A, Bhattacharya S, Acharya R (2005) Spatio-temporal dynamics of genomic organization and function in the mammalian cell nucleus. Adv Enzyme Regul 45:17–26
Bhattacharya S, Acharya R, Pliss A, Malyavantham KS, Berezney R (2005) Automated matching of genomic structures in microscopic images of living cells using an information theoretic approach. Conf Proc IEEE Eng Med Biol Soc 6:6425–6428
Bickmore WA, Mahy NL, Chambeyron S (2004) Do higher-order chromatin structure and nuclear reorganization play a role in regulating Hox gene expression during development? Cold Spring Harbor Symp Quant Biol 69:251–257
Boisvert FM, Hendzel MJ, Bazett-Jones DP (2000) Promyelocytic leukemia (PML) nuclear bodies are protein structures that do not accumulate RNA. J Cell Biol 148:283–292
Bolte S, Cordelieres FP (2006) A guided tour into subcellular colocalization analysis in light microscopy. J Microsc 224:213–232
Bregman DB, Du L, van der Zee S, Warren SL (1995) Transcription-dependent redistribution of the large subunit of RNA polymerase II to discrete nuclear domains. J Cell Biol 129:287–298
Chakalova L, Debrand E, Mitchell JA, Osborne CS, Fraser P (2005) Replication and transcription: shaping the landscape of the genome. Nat Rev 6:669–677
Chambeyron S, Bickmore WA (2004a) Chromatin decondensation and nuclear reorganization of the HoxB locus upon induction of transcription. Genes Dev 18:1119–1130
Chambeyron S, Bickmore WA (2004b) Does looping and clustering in the nucleus regulate gene expression? Curr Opin Cell Biol 16:256–262
Chubb JR, Trcek T, Shenoy SM, Singer RH (2006) Transcriptional pulsing of a developmental gene. Curr Biol 16:1018–1025
Cmarko D, Verschure PJ, Martin TE, Dahmus ME, Krause S, Fu XD, van Driel R, Fakan S (1999) Ultrastructural analysis of transcription and splicing in the cell nucleus after bromo-UTP microinjection. Mol Biol Cell 10:211–223
Cook P (1998) Duplicating a tangled genome. Science 281:1466–1467
Dehghani H, Dellaire G, Bazett-Jones DP (2005) Organization of chromatin in the interphase mammalian cell. Micron 36:95–108
Elbi C, Misteli T, Hager GL (2002) Recruitment of dioxin receptor to active transcription sites. Mol Biol Cell 13:2001–2015
Fakan S (1971) Inhibition of nucleolar RNP synthesis by cycloheximide as studied by high resolution radioautography. J Ultrastruct Res 34:586–596
Fraser P (2006) Transcriptional control thrown for a loop. Curr Opin Genet Dev 16:490–495
Gilbert DM (2002) Replication timing and transcriptional control: beyond cause and effect. Curr Opin Cell Biol 14:377–383
Goldman MA, Holmquist GP, Gray MC, Caston LA, Nag A (1984) Replication timing of genes and middle repetitive sequences. Science 224:686–692
Goren A, Cedar H (2003) Replicating by the clock. Nat Rev Mol Cell Biol 4:25–32
Grande MA, van der Kraan I, de Jong L, van Driel R (1997) Nuclear distribution of transcription factors in relation to sites of transcription and RNA polymerase II. J Cell Sci 110(Pt 15):1781–1791
Harrington KS, Javed A, Drissi H, McNeil S, Lian JB, Stein JL, Van Wijnen AJ, Wang YL, Stein GS (2002) Transcription factors RUNX1/AML1 and RUNX2/Cbfa1 dynamically associate with stationary subnuclear domains. J Cell Sci 115:4167–4176
Hatton KS, Dhar V, Brown EH, Iqbal MA, Stuart S, Didamo VT, Schildkraut CL (1988) Replication program of active and inactive multigene families in mammalian cells. Mol Cell Biol 8:2149–2158
He S, Davie JR (2006) Sp1 and Sp3 foci distribution throughout mitosis. J Cell Sci 119:1063–1070
He S, Sun JM, Li L, Davie JR (2005) Differential intranuclear organization of transcription factors Sp1 and Sp3. Mol Biol Cell 16:4073–4083
Iborra FJ, Pombo A, Jackson DA, Cook PR (1996) Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J Cell Sci 109(Pt 6):1427–1436
Jackson DA, Pombo A (1998) Replicon clusters are stable units of chromosome structure: evidence that nuclear organization contributes to the efficient activation and propagation of S phase in human cells. J Cell Biol 140:1285–1295
Jackson DA, Hassan AB, Errington RJ, Cook PR (1993) Visualization of focal sites of transcription within human nuclei. EMBO J 12:1059–1065
Jackson DA, Iborra FJ, Manders EM, Cook PR (1998) Numbers and organization of RNA polymerases, nascent transcripts, and transcription units in HeLa nuclei. Mol Biol Cell 9:1523–1536
Jackson DA, Pombo A, Iborra F (2000) The balance sheet for transcription: an analysis of nuclear RNA metabolism in mammalian cells. FASEB J 14:242–254
Lanctot C, Cheutin T, Cremer M, Cavalli G, Cremer T (2007) Dynamic genome architecture in the nuclear space: regulation of gene expression in three dimensions. Nat Rev 8:104–115
Levsky JM, Singer RH (2003) Gene expression and the myth of the average cell. Trends Cell Biol 13:4–6
Levsky JM, Shenoy SM, Pezo RC, Singer RH (2002) Single-cell gene expression profiling. Science 297:836–840
Levsky JM, Shenoy SM, Chubb JR, Hall CB, Capodieci P, Singer RH (2007) The spatial order of transcription in mammalian cells. J Cell Biochem 102:609–617
Lian JB, Stein GS, Javed A, van Wijnen AJ, Stein JL, Montecino M, Hassan MQ, Gaur T, Lengner CJ, Young DW (2006) Networks and hubs for the transcriptional control of osteoblastogenesis. Rev Endocr Metab Disord 7:1–16
Ling JQ, Li T, Hu JF, Vu TH, Chen HL, Qiu XW, Cherry AM, Hoffman AR (2006) CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312:269–272
Ma H, Samarabandu J, Devdhar RS, Acharya R, Cheng PC, Meng C, Berezney R (1998) Spatial and temporal dynamics of DNA replication sites in mammalian cells. J Cell Biol 143:1415–1425
MacAlpine DM, Rodriguez HK, Bell SP (2004) Coordination of replication and transcription along a Drosophila chromosome. Genes Dev 18:3094–3105
Mahy NL, Perry PE, Bickmore WA (2002a) Gene density and transcription influence the localization of chromatin outside of chromosome territories detectable by FISH. J Cell Biol 159:753–763
Mahy NL, Perry PE, Gilchrist S, Baldock RA, Bickmore WA (2002b) Spatial organization of active and inactive genes and noncoding DNA within chromosome territories. J Cell Biol 157:579–589
McManus KJ, Stephens DA, Adams NM, Islam SA, Freemont PS, Hendzel MJ (2006) The transcriptional regulator CBP has defined spatial associations within interphase nuclei. PLoS Comput Biol 2:e139
McNairn AJ, Gilbert DM (2003) Epigenomic replication: linking epigenetics to DNA replication. Bioessays 25:647–656
Mitchell JA, Fraser P (2008) Transcription factories are nuclear subcompartments that remain in the absence of transcription. Genes Dev 22:20–25
Mortillaro MJ, Blencowe BJ, Wei X, Nakayasu H, Du L, Warren SL, Sharp PA, Berezney R (1996) A hyperphosphorylated form of the large subunit of RNA polymerase II is associated with splicing complexes and the nuclear matrix. Proc Natl Acad Sci U S A 93:8253–8257
Nakamura H, Morita T, Sato C (1986) Structural organizations of replicon domains during DNA synthetic phase in the mammalian nucleus. Exp Cell Res 165:291–297
Nakayasu H, Berezney R (1989) Mapping replicational sites in the eukaryotic cell nucleus. J Cell Biol 108:1–11
Nunez E, Kwon YS, Hutt KR, Hu Q, Cardamone MD, Ohgi KA, Garcia-Bassets I, Rose DW, Glass CK, Rosenfeld MG, Fu XD (2008) Nuclear receptor-enhanced transcription requires motor- and LSD1-dependent gene networking in interchromatin granules. Cell 132:996–1010
Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, Fraser P (2004) Active genes dynamically colocalize to shared sites of ongoing transcription. Nat Genet 36:1065–1071
Patturajan M, Schulte RJ, Sefton BM, Berezney R, Vincent M, Bensaude O, Warren SL, Corden JL (1998) Growth-related changes in phosphorylation of yeast RNA polymerase II. J Biol Chem 273:4689–4694
Pombo A, Jones E, Iborra FJ, Kimura H, Sugaya K, Cook PR, Jackson DA (2000) Specialized transcription factories within mammalian nuclei. Crit Rev Eukaryot Gene Expr 10:21–29
Ragoczy T, Telling A, Sawado T, Groudine M, Kosak ST (2003) A genetic analysis of chromosome territory looping: diverse roles for distal regulatory elements. Chromosome Res 11:513–525
Raj A, Peskin CS, Tranchina D, Vargas DY, Tyagi S (2006) Stochastic mRNA synthesis in mammalian cells. PLoS Biol 4:e309
Ross IL, Browne CM, Hume DA (1994) Transcription of individual genes in eukaryotic cells occurs randomly and infrequently. Immunol Cell Biol 72:177–185
Sadoni N, Cardoso MC, Stelzer EH, Leonhardt H, Zink D (2004) Stable chromosomal units determine the spatial and temporal organization of DNA replication. J Cell Sci 117:5353–5365
Saltman LH, Javed A, Ribadeneyra J, Hussain S, Young DW, Osdoby P, Amcheslavsky A, van Wijnen AJ, Stein JL, Stein GS, Lian JB, Bar-Shavit Z (2005) Organization of transcriptional regulatory machinery in osteoclast nuclei: compartmentalization of Runx1. J Cell Physiol 204:871–880
Samarabandu J, Ma H, Acharya R, Cheng PC, Berezney R (1995) Image analysis techniques for visualizing the spatial organization of DNA replication sites in the mammalian cell nucleus using multi-channel confocal microscopy. SPIE 2434:370–375
Schubeler D, Scalzo D, Kooperberg C, van Steensel B, Delrow J, Groudine M (2002) Genome-wide DNA replication profile for Drosophila melanogaster: a link between transcription and replication timing. Nat Genet 32:438–442
Schwaiger M, Schubeler D (2006) A question of timing: emerging links between transcription and replication. Curr Opin Genet Dev 16:177–183
Shav-Tal Y, Darzacq X, Singer RH (2006) Gene expression within a dynamic nuclear landscape. EMBO J 25:3469–3479
Sparvoli E, Levi M, Rossi E (1994) Replicon clusters may form structurally stable complexes of chromatin and chromosomes. J Cell Sci 107(Pt 11):3097–3103
Spector DL (1993) Macromolecular domains within the cell nucleus. Annu Rev Cell Biol 9:265–315
Stein GS, Zaidi SK, Braastad CD, Montecino M, van Wijnen AJ, Choi JY, Stein JL, Lian JB, Javed A (2003) Functional architecture of the nucleus: organizing the regulatory machinery for gene expression, replication and repair. Trends Cell Biol 13:584–592
Stein GS, Lian JB, Stein JL, van Wijnen AJ, Javed A, Montecino M, Zaidi SK, Young DW, Choi JY, Pratap J (2005) Combinatorial organization of the transcriptional regulatory machinery in biological control and cancer. Adv Enzyme Regul 45:136–154
Stein GS, Lian JB, van Wijnen AJ, Stein JL, Javed A, Montecino M, Choi JY, Vradii D, Zaidi SK, Pratap J, Young D (2007) Organization of transcriptional regulatory machinery in nuclear microenvironments: implications for biological control and cancer. Adv Enzyme Regul 47:242–250
Tumbar T, Belmont AS (2001) Interphase movements of a DNA chromosome region modulated by VP16 transcriptional activator. Nat Cell Biol 3:134–139
van Steensel B, van Binnendijk EP, Hornsby CD, van der Voort HT, Krozowski ZS, de Kloet ER, van Driel R (1996) Partial colocalization of glucocorticoid and mineralocorticoid receptors in discrete compartments in nuclei of rat hippocampus neurons. J Cell Sci 109(Pt 4):787–792
Volpi EV, Chevret E, Jones T, Vatcheva R, Williamson J, Beck S, Campbell RD, Goldsworthy M, Powis SH, Ragoussis J, Trowsdale J, Sheer D (2000) Large-scale chromatin organization of the major histocompatibility complex and other regions of human chromosome 6 and its response to interferon in interphase nuclei. J Cell Sci 113(Pt 9):1565–1576
Wansink DG, Schul W, van der Kraan I, van Steensel B, van Driel R, de Jong L (1993) Fluorescent labeling of nascent RNA reveals transcription by RNA polymerase II in domains scattered throughout the nucleus. J Cell Biol 122:283–293
Wansink DG, Manders EE, van der Kraan I, Aten JA, van Driel R, de Jong L (1994) RNA polymerase II transcription is concentrated outside replication domains throughout S-phase. J Cell Sci 107(Pt 6):1449–1456
Wansink DG, Sibon OC, Cremers FF, van Driel R, de Jong L (1996) Ultrastructural localization of active genes in nuclei of A431 cells. J Cell Biochem 62:10–18
Warren SL, Landolfi AS, Curtis C, Morrow JS (1992) Cytostellin: a novel, highly conserved protein that undergoes continuous redistribution during the cell cycle. J Cell Sci 103(Pt 2):381–388
Wei X, Samarabandu J, Devdhar RS, Siegel AJ, Acharya R, Berezney R (1998) Segregation of transcription and replication sites into higher order domains. Science 281:1502–1506
Wei X, Somanathan S, Samarabandu J, Berezney R (1999) Three-dimensional visualization of transcription sites and their association with splicing factor-rich nuclear speckles. J Cell Biol 146:543–558
West AG, Fraser P (2005) Remote control of gene transcription. Hum Mol Genet 14 Spec No 1:R101–111
Woodfine K, Fiegler H, Beare DM, Collins JE, McCann OT, Young BD, Debernardi S, Mott R, Dunham I, Carter NP (2004) Replication timing of the human genome. Hum Mol Genet 13:191–202
Zaidi SK, Young DW, Choi JY, Pratap J, Javed A, Montecino M, Stein JL, van Wijnen AJ, Lian JB, Stein GS (2005) The dynamic organization of gene-regulatory machinery in nuclear microenvironments. EMBO Rep 6:128–133
Zaidi SK, Javed A, Pratap J, Schroeder TMJJW, Lian JB, van Wijnen AJ, Stein GS, Stein JL (2006) Alterations in intranuclear localization of Runx2 affect biological activity. J Cell Physiol 209:935–942
Zaidi SK, Young DW, Javed A, Pratap J, Montecino M, van Wijnen A, Lian JB, Stein JL, Stein GS (2007) Nuclear microenvironments in biological control and cancer. Nat Rev Cancer 7:454–463
Zeng C, Kim E, Warren SL, Berget SM (1997) Dynamic relocation of transcription and splicing factors dependent upon transcriptional activity. EMBO J 16:1401–1412
Acknowledgements
The authors are grateful to Dr. Daniela S. Dimitrova who first demonstrated in our laboratory the lack of colocalization between replication and transcription sites using standard epifluorescence microscopy. The authors thank Dr. George Mayers and Dr. Richard B. Bankert (University at Buffalo, NY) for providing anti-BrdU mouse monoclonal antibodies. These studies were supported by the National Institutes of Health (NIH) grant GM 072131 awarded to R.B.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by E.A. Nigg
Electronic supplementary material
Below is the link to the electronic supplementary material.
S1
Proximity analysis of 0.5 μm Fluoresbrite beads. Fluoresbrite beads of 0.5 μm diameter, which could fluoresce in both red and green colors, were imaged and analyzed with our segmentation and proximity programs. Including the sub-pixel error associated with CCD imaging, ∼85% of the beads were at a distance of 1 pixel to their nearest neighbor. Below is the proximity plot which describes their spatial association. (60.5 KB)
S2
Line profile analysis of B3 and ARNA3 antibodies. Comparison of the staining patterns produced by B3 (green, specific for hyperphosphorylated form of the RNA polymerase II large subunit) and ARNA3 (red, specific for total RNA polymerase II large subunit) antibodies. Line profile analysis of the merged image and two enlarged portions A and B, indicates the overlap between the stained patterns in the red and green channels. (153 KB)
Rights and permissions
About this article
Cite this article
Malyavantham, K.S., Bhattacharya, S., Alonso, W.D. et al. Spatio-temporal dynamics of replication and transcription sites in the mammalian cell nucleus. Chromosoma 117, 553–567 (2008). https://doi.org/10.1007/s00412-008-0172-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00412-008-0172-6