Abstract
Purpose
To evaluate the performance of chromosomal microarray analysis (CMA) in fetuses with nuchal translucency (NT) > 95th percentile. Secondary objectives were to analyze these results according to NT thickness, below or above 3.5 mm, and those without associated anomalies.
Methods
This observational single-cohort study was conducted between 2015 and 2018 in fetuses with NT > 95th percentile. Following an invasive test, quantitative fluorescence-polymerase chain reaction (QF-PCR) was performed, and if normal, CMA was performed. Pathogenic copy number variants (CNVs), non-reported pathogenic CNV, pathogenic autosomal recessive variants and variants of unknown significance (VUS) were analysed.
Results
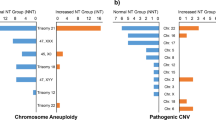
One-hundred and sixty-two fetuses with NT > 95th percentile, normal QF-PCR and CMA were included. Amongst 128 fetuses with NT between the 95th percentile and 3.5 mm, one (0.8%) had a pathogenic CNV, four (3.1%) had non-reported pathogenic CNV, one (0.8%) had pathogenic autosomal recessive variant and 13 (10.2%) had VUS. Amongst 34 fetuses with NT ≥ 3.5 mm, four (11.8%) had pathogenic CNV, one (2.9%) had non-reported pathogenic CNV, one (2.9%) had pathogenic autosomal recessive variant and four (11.8%) had VUS. Four in 162 (2.5%) fetuses had CNVs at the chromosome 16p13.11 region. Amongst 154 fetuses without structural abnormalities and normal QF-PCR, three (1.9%) had a pathogenic CNV, 5 (3.2%) had non-reported pathogenic CNV, one (0.6%) autosomal recessive pathogenic CNV and 16 (10.4%) had VUS.
Conclusion
Pathogenic CNVs were found in 1% of fetuses with an NT thickness between the 95th percentile and 3.5 mm and in 12% of fetuses with NT ≥ 3.5 mm. CNVs were found at the 16p13.11 region in 2.5% of cases.

Similar content being viewed by others
Abbreviations
- NT:
-
Nuchal translucency
- CGH:
-
Comparative genomic hybridization
- CNV:
-
Copy number variant
- QF-PCR:
-
Quantitative fluorescence-polymerase chain reaction
- CMA:
-
Chromosomal microarray analysis
- VUS:
-
Variant of unknown significance
- SD:
-
Standard deviation
- CI:
-
Confidence interval
- cfDNA:
-
Cell-free DNA
References
Souka AP, Von Kaisenberg CS, Hyett JA et al (2005) Increased nuchal translucency with normal karyotype. Am J Obstet Gynecol 192:1005–1021. https://doi.org/10.1016/j.ajog.2004.12.093
Baer RJ, Norton ME, Shaw GM et al (2014) Risk of selected structural abnormalities in infants after increased nuchal translucency measurement. Am J Obstet Gynecol 211:675.e1–19. https://doi.org/10.1016/j.ajog.2014.06.025
Kagan KO, Sonek J, Kozlowski P (2022) Antenatal screening for chromosomal abnormalities. Arch Gynecol Obstet 305:825–835. https://doi.org/10.1007/s00404-022-06477-5
Hellmuth SG, Pedersen LH, Miltoft CB et al (2017) Increased nuchal translucency thickness and risk of neurodevelopmental disorders. Ultrasound Obstet Gynecol 49:592–598. https://doi.org/10.1002/uog.15961
Wapner RJ, Martin CL, Levy B et al (2012) Chromosomal microarray versus karyotyping for prenatal diagnosis. N Engl J Med 367:2175–2184. https://doi.org/10.1056/NEJMoa1203382
Hillman SC, Pretlove S, Coomarasamy A et al (2011) Additional information from array comparative genomic hybridization technology over conventional karyotyping in prenatal diagnosis: a systematic review and meta-analysis. Ultrasound Obstet Gynecol 37:6–14. https://doi.org/10.1002/uog.7754
de Wit MC, Srebniak MI, Govaerts LCP et al (2014) Additional value of prenatal genomic array testing in fetuses with isolated structural ultrasound abnormalities and a normal karyotype: a systematic review of the literature: Genomic microarray testing in fetuses with structural anomalies. Ultrasound Obstet Gynecol 43:139–146. https://doi.org/10.1002/uog.12575
Vestergaard EM, Christensen R, Petersen OB, Vogel I (2013) Prenatal diagnosis: array comparative genomic hybridization in fetuses with abnormal sonographic findings. Acta Obstet Gynecol Scand 92:762–768. https://doi.org/10.1111/aogs.12146
Egloff M, Hervé B, Quibel T et al (2018) Diagnostic yield of chromosomal microarray analysis in fetuses with isolated increased nuchal translucency: a French multicenter study. Ultrasound Obstet Gynecol 52:715–721. https://doi.org/10.1002/uog.18928
Ma Y, Pei Y, Yin C et al (2019) Subchromosomal anomalies in small for gestational-age fetuses and newborns. Arch Gynecol Obstet 300:633–639. https://doi.org/10.1007/s00404-019-05235-4
Grande M, Jansen FAR, Blumenfeld YJ et al (2015) Genomic microarray in fetuses with increased nuchal translucency and normal karyotype: a systematic review and meta-analysis. Ultrasound Obstet Gynecol 46:650–658. https://doi.org/10.1002/uog.14880
Snijders RJ, Noble P, Sebire N et al (1998) UK multicentre project on assessment of risk of trisomy 21 by maternal age and fetal nuchal-translucency thickness at 10–14 weeks of gestation. Fetal Medicine Foundation First Trimester Screening Group. Lancet 352:343–346. https://doi.org/10.1016/s0140-6736(97)11280-6
Kagan KO, Wright D, Valencia C et al (2008) Screening for trisomies 21, 18 and 13 by maternal age, fetal nuchal translucency, fetal heart rate, free beta-hCG and pregnancy-associated plasma protein-A. Hum Reprod 23:1968–1975. https://doi.org/10.1093/humrep/den224
Generalitat de Catalunya. Departamento de Salud (2008) Protocolo de diagnóstico prenatal de anomalías congénitas fetales. [Prenatal diagnosis protocol for fetal congenital anomalies], Primera edición. Direcció General de Salut Pública, Barcelona
Wright D, Kagan KO, Molina FS et al (2008) A mixture model of nuchal translucency thickness in screening for chromosomal defects. Ultrasound Obstet Gynecol 31:376–383. https://doi.org/10.1002/uog.5299
Nicolaides KH (2011) Screening for fetal aneuploidies at 11 to 13 weeks. Prenat Diagn 31:7–15. https://doi.org/10.1002/pd.2637
Verma R, Babu A (1995) Human chromosomes. Principles and techniques. McGraw-Hill, New York
Cirigliano V, Ejarque M, Cañadas MP et al (2001) Clinical application of multiplex quantitative fluorescent polymerase chain reaction (QF-PCR) for the rapid prenatal detection of common chromosome aneuploidies. Mol Hum Reprod 7:1001–1006. https://doi.org/10.1093/molehr/7.10.1001
Kearney HM, Thorland EC, Brown KK et al (2011) American College of Medical Genetics standards and guidelines for interpretation and reporting of postnatal constitutional copy number variants. Genet Med 13:680–685. https://doi.org/10.1097/GIM.0b013e3182217a3a
Riggs ER, Andersen EF, Cherry AM et al (2020) Technical standards for the interpretation and reporting of constitutional copy-number variants: a joint consensus recommendation of the American College of Medical Genetics and Genomics (ACMG) and the Clinical Genome Resource (ClinGen). Genet Med 22:245–257. https://doi.org/10.1038/s41436-019-0686-8
Vanakker O, Vilain C, Janssens K et al (2014) Implementation of genomic arrays in prenatal diagnosis: The Belgian approach to meet the challenges. Eur J Med Genet 57:151–156. https://doi.org/10.1016/j.ejmg.2014.02.002
Gardiner C, Wellesley D, Kilby MD et al (2015) Recommendations for the use of chromosome microarray in pregnancy. London
Su L, Huang H, An G et al (2019) Clinical application of chromosomal microarray analysis in fetuses with increased nuchal translucency and normal karyotype. Mol Genet Genomic Med. https://doi.org/10.1002/mgg3.811
Xue S, Yan H, Chen J et al (2020) Genetic examination for fetuses with increased fetal nuchal translucency by genomic technology. Cytogenet Genome Res 160:57–62. https://doi.org/10.1159/000506095
Hui L, Pynaker C, Bonacquisto L et al (2021) Reexamining the optimal nuchal translucency cutoff for diagnostic testing in the cell-free DNA and microarray era: results from the Victorian Perinatal Record Linkage study. Am J Obstet Gynecol 225:527.e1-527.e12. https://doi.org/10.1016/j.ajog.2021.03.050
Sagi-Dain L, Singer A, Ben Shachar S et al (2021) Risk of clinically significant chromosomal microarray analysis findings in fetuses with nuchal translucency from 3.0 mm through 3.4 mm. Obstet Gynecol 137:126–131. https://doi.org/10.1097/AOG.0000000000004195
Petersen OB, Smith E, Van Opstal D et al (2020) Nuchal translucency of 3.0-3.4 mm an indication for NIPT or microarray? Cohort analysis and literature review. Acta Obstet Gynecol Scand 99:765–774. https://doi.org/10.1111/aogs.13877
Johnson K, Kelley J, Saxton V et al (2017) Declining invasive prenatal diagnostic procedures: a comparison of tertiary hospital and national data from 2012 to 2015. Aust N Z J Obstet Gynaecol 57:152–156. https://doi.org/10.1111/ajo.12590
Kagan KO, Sonek J, Wagner P, Hoopmann M (2017) Principles of first trimester screening in the age of non-invasive prenatal diagnosis: screening for chromosomal abnormalities. Arch Gynecol Obstet 296:645–651. https://doi.org/10.1007/s00404-017-4459-9
Srebniak MI, Knapen MFCM, Polak M et al (2017) The influence of SNP-based chromosomal microarray and NIPT on the diagnostic yield in 10,000 fetuses with and without fetal ultrasound anomalies. Hum Mutat 38:880–888. https://doi.org/10.1002/humu.23232
Konialis C, Pangalos C (2015) Dilemmas in prenatal chromosomal diagnosis revealed through a single center’s 30 years’ experience and 90,000 cases. Fetal Diagn Ther 38:218–232. https://doi.org/10.1159/000368604
Sotiriadis A, Papoulidis I, Siomou E et al (2017) Non-invasive prenatal screening versus prenatal diagnosis by array comparative genomic hybridization: a comparative retrospective study. Prenat Diagn 37:583–592. https://doi.org/10.1002/pd.5051
Muys J, Blaumeiser B, Jacquemyn Y et al (2018) The Belgian MicroArray Prenatal (BEMAPRE) database: a systematic nationwide repository of fetal genomic aberrations. Prenat Diagn 38:1120–1128. https://doi.org/10.1002/pd.5373
Law LW, Lau TK, Fung TY et al (2009) De novo 16p13.11 microdeletion identified by high-resolution array CGH in a fetus with increased nuchal translucency. BJOG 116:339–343. https://doi.org/10.1111/j.1471-0528.2008.01948.x
Paciorkowski AR, Keppler-Noreuil K, Robinson L et al (2013) Deletion 16p13.11 uncovers NDE1 mutations on the non-deleted homolog and extends the spectrum of severe microcephaly to include fetal brain disruption. Am J Med Genet A 161A:1523–1530. https://doi.org/10.1002/ajmg.a.35969
Tropeano M, Ahn JW, Dobson RJB et al (2013) Male-biased autosomal effect of 16p13.11 copy number variation in neurodevelopmental disorders. PLoS ONE 8:e61365. https://doi.org/10.1371/journal.pone.0061365
Maya I, Yacobson S, Kahana S et al (2017) Cut-off value of nuchal translucency as indication for chromosomal microarray analysis: NT and CMA. Ultrasound Obstet Gynecol 50:332–335. https://doi.org/10.1002/uog.17421
Maya I, Basel-Salmon L, Singer A, Sagi-Dain L (2020) High-frequency low-penetrance copy-number variant classification: should we revise the existing guidelines? Genet Med 22:1276–1277. https://doi.org/10.1038/s41436-020-0795-4
Funding
The authors declare that no funds, grants, or other support were received during the preparation of this manuscript.
Author information
Authors and Affiliations
Contributions
ECC: Protocol development, Data collection or management, Data analysis, Manuscript writing/editing. MAS-D: Protocol development, Data collection or management, Data analysis, Manuscript writing/editing. IC: Data collection or management, Manuscript writing/editing. MTH: Data collection or management, Manuscript writing/editing. MA-G: Data collection or management, Manuscript writing/editing. CR: Data collection or management, Manuscript writing/editing. NM: Data collection or management, Data analysis, Manuscript writing/editing. APR: Data collection or management, Manuscript writing/editing. NCS: Data collection or management, Manuscript writing/editing. CMV: Data collection or management, Manuscript writing/editing. EC: Manuscript writing/editing.
Corresponding author
Ethics declarations
Conflict of interest
The authors have no relevant financial or non-financial interests to disclose.
Ethical approval
This study was performed in line with the principles of the Declaration of Helsinki. The study was approved by the institutional research ethics committee of Vall d'Hebron University Hospital (CEIC-VHIR), PR(AMI)408/2019 on November 22nd 2019.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Coello-Cahuao, E., Sánchez-Durán, M.Á., Calero, I. et al. Array study in fetuses with nuchal translucency above the 95th percentile: a 4-year observational single-centre study. Arch Gynecol Obstet 307, 285–292 (2023). https://doi.org/10.1007/s00404-022-06564-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-022-06564-7