Abstract
Purpose
This study aimed to investigate the gut microbiome composition in pregnant women with digestive diseases to analyze the relationships between the microflora changes and digestive diseases during pregnancy.
Methods
Fecal samples obtained from 71 pregnant women [six acute fatty liver (AF group), 21 constipation (C group), 24 excessive vomiting (V group) and 20 normal pregnancy (CP group)] and 26 non-pregnant (NP group) women were subjected to 16 s rRNA sequencing. Differential analysis of intestinal flora at the genera level was performed.
Results
The relative abundance of Coprobacillus, Acinetobacter, Enterococcus, Weissella and Lysinibacillus was increased in the digestive diseases (AF, C and V) groups compared with CP group, whereas that of five common genera, including Terrisporobacter, Dysgonomonas, Adlercreutzia, Fusicatenibacter and Blautia, was decreased in digestive diseases groups. Additionally, in digestive diseases (AF, C and V) groups, the abundance of 13 common genera, such as Carnobacterium, Coprobacillus and Psychrobacter, was higher than NP group, whereas that of 27 common genera, such as Blautia and Terrisporobacter, was lower than NP group. About 69 genera were differentially abundant between AF and C groups; two genera (Aerococcus and Senegalimassilia) were identified between AF and V groups; moreover, total 63 genera were obtained between C and V groups.
Conclusion
Our data revealed that the abundance of Acinetobacter, Enterococci, Paenibacillus, Blautia and Collinsella might be associated with the digestive diseases during pregnancy. These findings further supported the idea that targeting the gut microbiota could be a new prevention or therapeutic approach for improving digestive diseases during pregnancy.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
During pregnancy, the pregnant woman usually undergoes significant physiological changes so as to accommodate and nurture the developing fetus, and these changes can result in a diversity of symptoms [1, 2]. It has been reported that pregnancy is a challenging period for the gastroenterologist, and multiple gastrointestinal complaints such as nausea, vomiting, gastroesophageal reflux, heartburn and constipation may occur [3]. The prevalence of nausea ranges from 50 to 80% and of vomiting is 50% [4]. Hyperemesis gravidarum is a severe form of nausea and vomiting, which affects about 1.2% of pregnant women [5].
In addition to gastrointestinal disorders, pregnancy is also associated with increased liver metabolism, and 3–5% of pregnancies present abnormal liver functions [6]. Liver disease during pregnancy is non-negligible, which may be life threatening for mother, fetus and subsequently the surviving child [7]. Acute fatty liver of pregnancy is an uncommon liver disease that occurs almost exclusively in the third trimester [8]. The reported incidence values for this disease are 1:7000–15,000 pregnancies [9, 10]. Acute fatty liver of pregnancy is a potentially fatal disease needing early diagnosis and intervention to prevent maternal and fetal mortality [6].
Growing evidences have shown that intestinal gut microbiota, a robust ecosystem inhabited by nearly 100 trillion bacteria, has an influence on intestinal physiological function and is involved in the life activities [11, 12]. For instance, chronic constipation, a prevalent functional gastrointestinal disorder, is demonstrated to be accompanied with intestinal dysbiosis [13]. Boursier et al. [14] recently reported that the severity of nonalcoholic fatty liver disease was implicated in gut dysbiosis and changes in metabolic function of the gut microbiota. However, there is presently little research about the changes in intestinal microbial communities in pregnant women with digestive diseases, and about how these changes affect the digestive system of patients.
In this study, we investigated the gut microbiome composition in fecal samples from pregnant women with digestive diseases, to analyze the relationships between the microflora changes and digestive diseases during pregnancy. The results may provide certain theoretical basis for safe, noninvasive diagnosis, treatment and intervention of digestive diseases during pregnancy.
Materials and methods
Sample collection
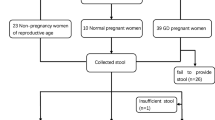
A total of 71 pregnant and 26 non-pregnant (NP group) women were involved in this study. Among the pregnant women, six cases had acute fatty liver (AF group), 21 cases had constipation (C group), 24 cases had excessive vomiting (V group) and 20 cases had normal pregnancy (CP group). The detailed information is listed in supplementary Table 1. Fecal samples were obtained from 97 women and stored at − 80 °C within 2 h.
This study was approved by the medical ethics committee of Qilu Hospital of Shandong University. All participants had given the informed written consent prior to their participation.
Genome DNA extraction and amplicon generation
Total genome DNA was extracted from fecal samples using cetyltrimethylammonium bromide and sodium dodecyl sulfate method. Following concentration and purity monitoring on 1% agarose gels, DNA was diluted with sterile water to 1 ng/μL. The 16S rRNA amplicons covering variable region V4 were amplified using specific primer (16S V4: 515F-806R). PCR reaction was carried out with Phusion® High-Fidelity PCR Master Mix (New England Biolabs, Beverly, MA, USA). The PCR products were quantified with electrophoresis on 2% agarose gel, mixed in equidensity ratios and then purified by Qiagen Gel Extraction Kit (Qiagen, Hilden, Germany).
Library preparation and sequencing
Library was prepared using TruSeq® DNA PCR-Free Sample Preparation Kit (Illumina, San Diego, CA, USA) followed by quality evaluation on the Qubit@ 2.0 Fluorometer and Agilent Bioanalyzer 2100 system. Library was finally sequenced on the Illumina HiSeq 2500 platform. The 250 bp paired-end reads were produced.
Paired-end reads assembly and quality control
Paired-end reads were merged using FLASH (V1.2.7) [15]. The raw tags were subjected to quality filtering to select the clean tags with high quality [16] using QIIME (V1.7.0) [17]. The clean tags were then mapped to the reference database (Gold database, http://drive5.com/uchime/uchime_download.html) using UCHIME algorithm [18], followed by removal of the chimera sequences [19] to identify the effective tags.
Operational taxonomic unit (OTU) clustering and annotation
Sequences were analyzed using Uparse software (Uparse V7.0.1001) [20]. Sequences with similarity of ≥ 97% were assigned to the same OTUs. Representative sequences for each OTU were selected for taxonomic information annotation using Mothur method and the SSUrRNA database [21] in SILVA [22].
Alpha diversity analysis
The abundance of OTUs was normalized using a standard of sequence number corresponding to the sample with the least sequence. The complexity of species diversity in the sample was analyzed using alpha diversity via seven indices, including Chao1, ACE, Shannon, Simpson, Goods_coverage, Observed_species and PD_whole_tree. The Chao1 and ACE estimator were used to identify the community richness. Shannon and Simpson indices were used to identify community diversity. The Goods_coverage was used to characterize the sequencing depth. PD_whole_tree index was used to identify phylogenetic diversity.
Beta diversity analysis
The differences of samples in species complexity were evaluated using beta diversity analysis. Beta diversity on unweighted UniFrac was calculated using QIIME to construct unweighted pair-group method with arithmetic means (UPGMA) sample clustering tree. Clustering analysis was performed by principal component analysis (PCA) using ade4 and ggplot2 packages in R software (V2.15.3). Principal coordinate analysis (PCoA) was conducted to get principal coordinates from complex multidimensional data, which were displayed by stat, WGCNA and ggplot2 packages in R software (V2.15.3).
Differential analysis of intestinal flora at the genera level
OTUs were filtered according to the OTU abundance using QIIME [17] tool, and the OTUs with abundance less than 1/10000 (0.0001) were deleted in this analysis. Then, the remaining OTUs were further screened based on their genus information, and the OTUs that did not annotate to the genus level were removed. Finally, 129 OTUs were obtained. The difference analysis was performed with the edgeR package in R. The OTU screening standard for significant difference was p value < 0.05, and false discovery rate (FDR) was used for p value adjustment.
Results
Sequencing data
After sequencing, the raw reads for all samples ranged from 54,075 to 99,209. The Q20 was more than 98% and Q30 was more than 97% for all samples. After quality control, the effective tags ranged from 80.72 to 95.17%.
OTU annotation
The top 10 maximum abundance of species in each group in genus level is shown in Fig. 1a. Bacteroides had the highest relative abundances in V, AF, CP and NP groups, and Faecalibacterium had the highest relative abundance in C group. In order to compare the species content in different groups of samples, the top 35 genera were selected for clustering analysis based on the species annotation and abundance information of samples at genus level. As shown in Fig. 1b, in the C group, Faecalibacterium, Bacteroides and Subdoligranulum were the dominant genera; in the V group, Bacteroides, Faecalibacterium and unidentified Enterobacteriaceae were the primary genera; in the AF group, Bacteroides, unidentified Enterobacteriaceae and Faecalibacterium were the dominant genera; in the CP group, Bacteroides, Faecalibacterium and Blautia were of higher abundance; meanwhile, in the NP group, the most abundant sequences were related to Bacteroides, Blautia and Faecalibacterium.
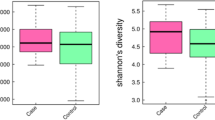
Alpha diversity analysis
The alpha diversity indexes, including observed species, Chao1, Shannon, Simpson, ACE, Goods_coverage and PD_whole_tree, for five groups are shown in Table 1. Generally, the community richness, community diversity and phylogenetic diversity in AF and V groups were higher than those in the other three groups. The rarefaction curve showed that the curve of each group tended to flatten, indicating that increasing sequencing depths did not help to discover new OTUs, which was in accordance with the sequencing depth index (Goods_coverage) as shown in Table 1.
Beta diversity analysis
PCoA analysis (unweighted UniFrac distance) showed that the samples in the NP group tended to cluster together, and samples in AF and V groups tended to cluster together (PC1, which explained 36.64% of variation in the community) (Fig. 2a). PCA revealed that the community structure was distinct between NP and the other groups (PC1, which explained 10.33% of variation in the community). Furthermore, the UPGMA clustering tree in phylum level was generated (Fig. 2b). In accordance with PCoA results, AF and V groups were clustered together.
a Results of PCoA. The x-axis represents one principal component, the y-axis represents another principal component and the percentage represents the contribution of the principal component to the sample difference. Each node represents a sample, and the samples from the same group are represented by the same color. b UPGMA clustering tree in phylum level
Differential analysis of intestinal flora at the genera level
The differential microorganisms in digestive diseases (AF, C and V) groups versus CP group, digestive diseases (AF, C and V) groups versus NP group and among digestive diseases groups (AF, C and V) were analyzed. The abundance of 13, 50 and 34 genera was increased in AF, V and C groups compared with CP group, respectively; inversely, a total of 5, 28 and 54 genera were reduced in AF, V and C groups compared with CP group. The heatmaps of three comparison groups are shown in Fig. 3a. In the three comparison groups, the abundance of five common genera, namely Coprobacillus, Acinetobacter, Enterococcus, Weissella and Lysinibacillus, was increased, while that of five common genera, namely Terrisporobacter, Dysgonomonas, Adlercreutzia, Fusicatenibacter and Blautia, was identified as decreasing (Fig. 3a, b).
a Visualized heatmaps of the differential microbiota (genus level) between acute fatty liver group (AF)/constipation group (C)/excessive vomiting (V group) and normal pregnancy group (CP). Red indicates high relative abundance, and green indicates low relative abundance. b Visualization results of intestinal flora differences in the intestinal diseases group (AF, C, V) versus the normal pregnancy group (CP). Red indicates the significantly increased genera in intestinal disease group compared with the normal pregnancy group. Blue indicates the significantly decreased genera in intestinal disease group compared with the normal pregnancy group
There were 31, 47 and 48 genera which were increased among digestive diseases (AF, C and V) groups compared with NP group, respectively; in addition, 32, 45 and 56 genera were decreased in the above three comparison groups. The heatmaps of three comparison groups are shown in Fig. 4a. A total of 13 common genera such as Carnobacterium, Coprobacillus, Caproiciproducens and Psychrobacter were observed to be increasing, while 27 common genera such as Novosphingobium, Dorea, Brevundimonas and Terrisporobacter were identified to be decreasing (Fig. 4a, b).
a Visualized heatmaps of the differential microbiota (genus level) between acute fatty liver group (AF)/constipation group (C)/excessive vomiting (V group) and non-pregnancy group (NP). Red indicates high relative abundance, and green indicates low relative abundance. b Visualization results of intestinal flora differences in the intestinal diseases group (AF, C, V) versus the normal pregnancy group (CP). Red indicates the significantly increased genera in intestinal disease group compared with the non-pregnancy group. Blue indicates the significantly decreased genera in intestinal disease group compared with the non-pregnancy group
Furthermore, in the AF group, the abundance of 38 genera was increased and 31 genera were decreased compared with C group. The relative abundances of Aerococcus and Senegalimassilia in AF group were reduced than V group. In the C group, an increased abundance of 23 genera and a decreased abundance of 40 genera were found than V group (Fig. 5).
Discussion
This study for the first time investigated the gut microbiome composition from pregnant women with digestive diseases by applying 16 s S rRNA gene sequencing. After analysis of the sequencing data, some differential genera were identified, which may serve as biomarkers of digestive diseases during pregnancy.
Acinetobacter is a gram-negative coccobacillus, which has emerged from a suspiciously pathogenic organism to an infectious agent important to hospitals around the world [23]. The species commonly cause nosocomial infections, but can also cause urinary tract and soft tissue infections [24]. Enterococci are gram-positive bacteria that usually inhabit the alimentary tract of humans [25]. Now enterococci have become common nosocomial pathogens, and enterococcal infections include surgical wound infection, hepatobiliary sepsis, urinary tract infections, bacteraemia and neonatal sepsis endocarditis [26]. Lysinibacillus, a ubiquitous gram-positive bacterium, is often regarded as environmental contaminants and rarely associated with human disease [27]. Recently, a case of sepsis due to bacteremia from Lysinibacillus and Paenibacillus was reported [28]. Given the pathogenicity of the three genera, we speculated that their increase in digestive disease groups compared with CP group may be implicated in the digestive diseases during pregnancy.
Blautia are gram-reaction-positive, non-motile bacteria that are obligate anaerobes, and most species of Blautia are isolated from the feces of humans and other mammals [29]. The Blautia coccoides is the dominant bacterium in the human intestine and accounts for approximately 20–30% of all bacteria in healthy person [30]. It has been reported that decreased relative abundance of the Blautia genus in the human gut is associated with malnutrition [31]. The gram-positive genus Collinsella contains three recognized species (Collinsella stercoris, C. aerofaciens and C. intestinalis), all of which are isolated from the feces of healthy human [32]. The relative abundance of Blautia and Collinsella decreased was related to digestive diseases during pregnancy, and Blautia and Collinsella might serve as novel biomarkers in diagnosis and treatment of digestive diseases.
We also compared the differential microorganisms among various digestive diseases groups. Interestingly, only two differential microorganisms were identified between AF and V groups, suggesting that there was no significant difference in intestinal flora between the AF group and V group. The underlying reason needs to be further explored. Members of Paenibacillus have been isolated from water, soil, diseased insect larvae and foods, presenting physiologically diverse characteristics [33]. Some Paenibacillus species, such as Paenibacillus alvei, P. polymyxa and P. macerans, are pathogenic, which have been isolated from patients suffering from sickle cell anemia, meningitis and wound infection [34]. In this study, the abundance of Paenibacillus was increased between AF versus V group and V versus C group, suggesting an increased expression trend from C to AF. We speculated that increased Paenibacillus in intestinal tract may indicate the severity levels of digestive diseases.
In conclusion, our study compared the differences between pregnancy with digestive diseases and normal pregnancy or non-pregnancy. The increase of Acinetobacter, Enterococci and Paenibacillus, and decrease of Blautia and Collinsella may be associated with the digestive diseases during pregnancy.
References
Soma-Pillay P, Catherine N-P, Tolppanen H, Mebazaa A (2016) Physiological changes in pregnancy. Cardiovasc J Afr 27(2):89
Gomes CF, Sousa M, Lourenço I, Martins D, Torres J (2018) Gastrointestinal diseases during pregnancy: what does the gastroenterologist need to know? Ann Gastroenterol 31(4):385
McCarthy FP, Lutomski JE, Greene RA (2014) Hyperemesis gravidarum: current perspectives. Int J Women’s Health 6:719
Matthews A, Haas DM, O’Mathúna DP, Dowswell T (2010) Interventions for nausea and vomiting in early pregnancy. Cochrane Database Syst Rev 8(9):CD007575
Einarson TR, Piwko C, Koren G (2013) Prevalence of nausea and vomiting of pregnancy in the USA: a meta-analysis. J Popul Ther Clin Pharmacol 20(2):e163
Hay JE (2008) Liver disease in pregnancy. Hepatology 47(3):1067–1076
Boregowda G, Shehata HA (2013) Gastrointestinal and liver disease in pregnancy. Best Pract Res Clin Obstet Gynaecol 27(6):835–853
Browning MF, Levy HL, Wilkins-Haug LE, Larson C, Shih VE (2006) Fetal fatty acid oxidation defects and maternal liver disease in pregnancy. Obstet Gynecol 107(1):115–120
Nelson DB, Yost NP, Cunningham FG (2013) Acute fatty liver of pregnancy: clinical outcomes and expected duration of recovery. Am J Obstet Gynecol 209(5):456.e1–456.e7
Allen AM, Kim WR, Larson JJ, Rosedahl JK, Yawn BP, McKeon K et al (2016) The epidemiology of liver diseases unique to pregnancy in a US community: a population-based study. Clin Gastroenterol Hepatol 14(2):287-94.e2
de Meij TG, de Groot EF, Eck A, Budding AE, Kneepkens CF, Benninga MA et al (2016) Characterization of microbiota in children with chronic functional constipation. PLoS ONE 11(10):e0164731
Simrén M, Barbara G, Flint HJ, Spiegel BM, Spiller RC, Vanner S et al (2013) Intestinal microbiota in functional bowel disorders: a Rome foundation report. Gut 62(1):159–176
Cao H, Liu X, An Y, Zhou G, Liu Y, Xu M et al (2017) Dysbiosis contributes to chronic constipation development via regulation of serotonin transporter in the intestine. Sci Rep 7(1):10322
Boursier J, Mueller O, Barret M, Machado M, Fizanne L, Araujo-Perez F et al (2016) The severity of nonalcoholic fatty liver disease is associated with gut dysbiosis and shift in the metabolic function of the gut microbiota. Hepatology 63(3):764–775
Magoä T, Salzberg SL (2011) FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27(21):2957–2963
Bokulich NA, Subramanian S, Faith JJ, Gevers D, Gordon JI, Knight R et al (2013) Quality-filtering vastly improves diversity estimates from Illumina amplicon sequencing. Nat Methods 10(1):57–59
Caporaso J, Kuczynski J, Stombaugh J, Bittinger K, Bushman F, Costello E et al (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7(5):335–336
Edgar RC, Haas BJ, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27(16):2194
Haas BJ, Gevers D, Earl AM, Feldgarden M, Ward DV, Giannoukos G et al (2011) Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res 21(3):494
Edgar RC (2013) UPARSE: highly accurate OTU sequences from microbial amplicon reads. Nat Methods 10(10):996–998
Quast C, Pruesse E, Yilmaz P, Gerken J, Schweer T, Yarza P et al (2013) The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res 41(Database issue):590–596
Wang Q, Garrity GM, Tiedje JM, Cole JR (2007) Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl Environ Microbiol 73(16):5261
Fournier PE, Richet H, Weinstein RA (2006) The epidemiology and control of Acinetobacter baumannii in health care facilities. Clin Infect Dis 42(5):692–699
Wong D, Nielsen TB, Bonomo RA, Pantapalangkoor P, Luna B, Spellberg B (2017) Clinical and pathophysiological overview of Acinetobacter infections: a century of challenges. Clin Microbiol Rev 30(1):409–447
Fisher K, Phillips C (2009) The ecology, epidemiology and virulence of Enterococcus. Microbiology 155(6):1749–1757
Poh C, Oh H, Tan A (2006) Epidemiology and clinical outcome of enterococcal bacteraemia in an acute care hospital. J Infect 52(5):383–386
Castagnola E, Fioredda F, Barretta M, Pescetto L, Garaventa A, Lanino E et al (2001) Bacillus sphaericus bacteraemia in children with cancer: case reports and literature review. J Hosp Infect 48(2):142–145
Wenzler E, Kamboj K, Balada-Llasat J-M (2015) Severe sepsis secondary to persistent Lysinibacillus sphaericus, Lysinibacillus fusiformis and Paenibacillus amylolyticus bacteremia. Int J Infect Dis 35:93–95
Liu C, Finegold SM, Song Y, Lawson PA (2008) Reclassification of Clostridium coccoides, Ruminococcus hansenii, Ruminococcus hydrogenotrophicus, Ruminococcus luti, Ruminococcus productus and Ruminococcus schinkii as Blautia coccoides gen. nov., comb. nov., Blautia hansenii comb. nov., Blautia hydrogenotrophica comb. nov., Blautia luti comb. nov., Blautia producta comb. nov., Blautia schinkii comb. nov. and description of Blautia wexlerae sp. nov., isolated from human faeces. Int J Syst Evol Microbiol 58(8):1896–1902
Matsuki T, Watanabe K, Fujimoto J, Takada T, Tanaka R (2004) Use of 16S rRNA gene-targeted group-specific primers for real-time PCR analysis of predominant bacteria in human feces. Appl Environ Microbiol 70(12):7220–7228
Million M, Diallo A, Raoult D (2017) Gut microbiota and malnutrition. Microb Pathog 106:127–138
Nagai F, Watanabe Y, Morotomi M (2010) Slackia piriformis sp. nov. and Collinsella tanakaei sp. nov., new members of the family Coriobacteriaceae, isolated from human faeces. Int J Syst Evol Microbiol 60(11):2639–2646
Daane L, Harjono I, Barns S, Launen L, Palleron N, Häggblom M (2002) PAH-degradation by Paenibacillus spp. and description of Paenibacillus naphthalenovorans sp. nov., a naphthalene-degrading bacterium from the rhizosphere of salt marsh plants. Int J Syst Evol Microbiol 52(1):131–139
Ko KS, Kim Y-S, Lee MY, Shin SY, Jung DS, Peck KR et al (2008) Paenibacillus konsidensis sp. nov., isolated from a patient. Int J Syst Evol Microbiol 58(9):2164–2168
Acknowledgements
This work was supported by the National Natural Science Foundation of China (No. 81700890) and Natural Science Foundation of Shandong Province (No. ZR2017BH115).
Author information
Authors and Affiliations
Contributions
YL, MJ and RJ conceptualized and designed the research. MJ, WL and XX were involved in acquisition of data. MJ, RJ and DL analyzed and interpreted the data. MJ and DL were involved in statistical analysis. MJ drafted the manuscript. YL revised the manuscript for important intellectual content.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflicts of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Jin, M., Li, D., Ji, R. et al. Changes in intestinal microflora in digestive tract diseases during pregnancy. Arch Gynecol Obstet 301, 243–249 (2020). https://doi.org/10.1007/s00404-019-05336-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00404-019-05336-0