Abstract
Introduction
Circulating tumour DNA (ctDNA) has emerged as a promising biomarker in various cancer types, including locally advanced rectal cancer (LARC), offering potential insights into disease progression, treatment response and recurrence. This review aims to comprehensively evaluate the utility of ctDNA as a prognostic biomarker in LARC.
Methods
PubMed, EMBASE and Web of Science were searched as part of our review. Studies investigating the utility of ctDNA in locally advanced rectal cancer (LARC) were assessed for eligibility. Quality assessment of included studies was performed using the Newcastle Ottawa Scale (NOS) risk of bias tool. Outcomes extracted included basic participant characteristics, ctDNA details and survival data. A meta-analysis was performed on eligible studies to determine pooled recurrence-free survival (RFS).
Results
Twenty-two studies involving 1676 participants were included in our analysis. Methodological quality categorised by the Newcastle Ottawa Scale was generally satisfactory across included studies. ctDNA detected at various time intervals was generally associated with poor outcomes across included studies. Meta-analysis demonstrated a pooled hazard ratio of 8.87 (95% CI 4.91–16.03) and 15.15 (95% CI 8.21–27.95), indicating an increased risk of recurrence with ctDNA positivity in the post-neoadjuvant and post-operative periods respectively.
Conclusion
Our systematic review provides evidence supporting the prognostic utility of ctDNA in patients with LARC, particularly in identifying patients at higher risk of disease recurrence in the post-neoadjuvant and post-operative periods.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Locally advanced rectal cancer (LARC) can cause significant challenges in terms of management [1]. Increasingly, patients are having total neoadjuvant treatment (TnT) [2], and the need for better indicators of complete clinical response, especially in borderline cases, is vital [3]. Circulating tumour DNA (ctDNA) has emerged as a promising biomarker in various cancer types, including LARC, offering potential insights into disease progression, treatment response and recurrence [4,5,6,7,8].
ctDNA refers to fragmented DNA shed by tumour cells into the bloodstream [9]. These fragments carry genetic alterations characteristic of the originating tumour, providing a non-invasive means of interrogating tumour biology [10]. The detection and analysis of ctDNA have garnered significant interest in cancer research due to its potential applications in diagnosis, prognostication and treatment monitoring [11]. ctDNA can be isolated from peripheral blood samples and analysed using various techniques, including next-generation sequencing (NGS), digital PCR (dPCR) and targeted amplicon-based assays [12].
NGS is a highly sensitive technique that allows for the comprehensive profiling of ctDNA, enabling the detection of a wide range of genetic alterations, including single nucleotide variants (SNVs), insertions and deletions (indels), copy number variations (CNVs) and structural rearrangements [13]. Conversely, dPCR quantifies the absolute number of target DNA molecules, allowing for precise measurement of ctDNA levels and increased cost-effectiveness when compared to NGS [14]. Alternative approaches include real-time PCR (qPCR), BEAMing and fragment analysis [15,16,17]. By profiling the genomic landscape of tumours through ctDNA analysis, clinicians can gain insights into tumour heterogeneity, clonal evolution and potential therapeutic targets, thereby facilitating personalised treatment approaches [18].
While several clinicopathological factors (MRI and endoscopic response) are currently used to stratify response to treatments in patients with LARC, they have limitations [19]. In addition, traditional prognostic factors such as tumour stage and histological grade can provide circumstantial value about potential disease behaviour and treatment response. There is a need to identify novel biomarkers that can complement existing prognostic tools and enhance risk stratification in LARC [20]. This systematic review aims to comprehensively evaluate the utility of ctDNA as a prognostic biomarker in LARC, exploring its potential to address the unmet clinical needs in this challenging disease context.
Methods
Study design and reporting guidelines
This is a systematic review and meta-analysis of retrospective and prospective cohort studies conducted in accordance with the Preferred Reporting Items for Systematic Reviews and Meta-Analyses (PRISMA) reporting guidelines [21]. Local institutional ethical approval was not required. All authors declare no conflicts of interest. This research received no external funding.
Search strategy
The following databases were searched as part of our systematic review process in April 2024; MEDLINE, PubMed, Embase and Web of Science. The following search strategy was used: (rectal OR rectum OR colorect*) AND (circulating tumour DNA OR ctDNA OR ct-DNA OR circulating free DNA OR cfDNA or cf-DNA). The search was completed on 5th April 2024. The grey literature was also searched as part of our study to identify any further ongoing works of literature, including theses and conference abstracts.
Eligibility criteria
Original studies investigating an association between ctDNA and treatment or oncological outcomes in patients with locally advanced rectal cancer were eligible for inclusion. Case reports, conference abstracts and review articles were excluded.
Data extraction and quality assessment
A database was established utilising the citation management software EndNote X9TM. Independent reviews of search outputs were conducted by two researchers (NOS and HCT). Initially, duplicate entries were eradicated, followed by a screening of study titles to gauge potential relevance. Subsequently, the abstracts of selected studies underwent assessment for eligibility based on predetermined inclusion/exclusion criteria. Excluded studies were categorised by reason within the database. Full texts of eligible abstracts were then scrutinised using identical criteria.
For efficient data extraction and storage, the Cochrane Collaboration’s screening and data extraction tool, Covidence, was employed [22]. Data collection was undertaken independently by two reviewers (NOS and HCT), encompassing study details, design, population, intervention, comparison groups and outcomes. Discrepancies between reviewers were resolved through open discussion, with final arbitration by the senior author (MK).
Potential biases in non-RCT studies were evaluated using the Newcastle-Ottawa Scale (NOS) risk of bias tool, with results tabulated accordingly [23]. This tool assesses studies across various categories, assigning stars to denote quality: 7 stars indicating “very good,” 5-6 stars for “good,” 3-4 stars for “satisfactory,” and 0–2 stars for “unsatisfactory.” Two reviewers (NOS and HCT) independently conducted critical appraisals, with a third reviewer (MK) arbitrating in cases of discordance.
Statistical analysis
Statistical analysis was conducted using Revman Statistical Software (Ver. 5 Copenhagen, Denmark). Generic inverse variance data were presented as hazard ratios (HR) alongside 95% confidence intervals (95% CI). Outcome measures, including mean with standard deviation and median with interquartile range, were documented. Only studies that provided hazard ratios with either confidence intervals or p-values were eligible for inclusion in the meta-analysis. When necessary, outcome variables (mean and SD) were estimated from the median and range using the formula described by Hozo et al. [24]. Heterogeneity was evaluated using I-squared statistics, with values exceeding 50% indicating significant heterogeneity. Statistical significance was defined as a p-value of less than 0.05. A random effects model was employed uniformly.
Systematic review registration
Our systematic review was registered on PROSPERO in April 2024 (ID: 533712).
Results
Search results
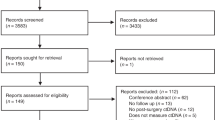
The previously outlined literature search yielded a total of 2123 results (Supplementary material S1). After eliminating 281 duplicates, 1842 studies underwent screening. Following the initial screening, 73 full abstracts were meticulously reviewed for eligibility, resulting in 42 being selected for full-text scrutiny. Among these 42 full texts, 22 studies met the eligibility criteria and were consequently included in our analysis [25,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45,46]. Notably, eight of the included studies provided adequate statistical data for incorporation into our quantitative analysis [30, 32,33,34, 36, 42, 44, 45]. A detailed depiction of the literature screening process can be found in Supplementary material S1.
Methodological characteristics and quality of studies
Among the 22 studies included, seventeen were conducted prospectively, four retrospectively, and one did not specify the study design. Regarding the study settings, ten were conducted in a single institute, nine in a multi-institutional setting, and three did not specify. Concerning the assessment of the risk of bias, eleven studies were rated as “very good”, ten as “good”, one as “satisfactory” and none as “unsatisfactory” according to the classification by the Newcastle Ottawa Scale. Supplementary material S2 provides a summary of our risk of bias assessment. The methodological characteristics of the included studies are outlined in Supplementary material S3.
Participant characteristics
The total number of participants across the included studies was 1676. All patients had a diagnosis of locally advanced rectal cancer. Baseline participant characteristics are outlined below in Table 1.
ctDNA details and study findings
ctDNA assay type, sequencing method and collection time points varied across included studies. Fourteen studies measured a mutation-specific panel, six measured cfDNA concentration and the remaining two measured promoter genes or multiple assays respectively. In regard to the sequencing method, next-generation sequencing (NGS) was the most commonly utilised method accounting for ten studies, followed by polymerase chain reaction (PCR) (n = 9) and direct fluorescent antibody (DFA) (n = 2). One study failed to report the sequencing method.
In terms of approach, eleven studies used an agnostic, ten used targeted and one study used both approaches. Agnostic approaches do not rely on prior knowledge of specific mutations but instead analyse the entire ctDNA for any alterations that may be present [47]. These approaches are valuable in settings where comprehensive genomic profiling is necessary, particularly for identifying novel or unexpected mutations. They are beneficial in research and clinical scenarios where a wide array of genetic alterations needs to be detected to inform treatment decisions or understand tumour heterogeneity [48]. Conversely, targeted approaches focus on detecting specific, known mutations that are of clinical interest [49]. They often use panels designed to target commonly mutated genes in particular cancers. Targeted approaches are highly relevant in clinical settings where specific genetic alterations are known to influence prognosis or guide targeted therapies [50]. They are typically more cost-effective and faster than agnostic approaches, making them suitable for routine clinical use, particularly when monitoring known mutations for treatment response or minimal residual disease. Both approaches have their place in the evaluation of ctDNA [47]. Agnostic methods provide a broad view of the genetic landscape, which can uncover new targets for therapy, while targeted methods allow for focused, efficient and cost-effective monitoring of known genetic alterations.
Tables 2 and 3 below outline ctDNA panel details, collection timepoints and main study findings.
Meta-analysis
Eight of the included studies provided sufficient statistical data to be included in our quantitative analysis [30, 32,33,34, 36, 42, 44, 45]. We investigated for an association between recurrence-free survival and the presence of ctDNA at several timepoints in a meta-analysis, the results of which are demonstrated in Fig. 1 below. The pooled hazard ratio for ctDNA presence after completion of neoadjuvant therapy when compared to patients who were not found to have detectable ctDNA was 8.87 (95% CI 4.91–16.03, p = < 0.0001)). Similarly, the pooled hazard ratio for ctDNA presence post-operatively compared to those without detectable ctDNA was 15.15 (95% CI 8.21–27.95, p = < 0.0001)). These results indicate an increased risk for recurrence in patients with LARC with detectable ctDNA in either the post-neoadjuvant therapy or post-operative periods.
Ongoing research
Several trials are ongoing to further determine the prognostic capabilities of ctDNA in patients with LARC. The DYNAMIC-RECTAL trial (ACTRN12617001560381) is a multi-centre randomised controlled phase 2 trial aiming to investigate the prognostic benefits of ctDNA detection post-operatively to guide the need for adjuvant treatment in patients with LARC. Eligible patients underwent neoadjuvant chemoradiation and total mesenteric excision, and the primary endpoint was adjuvant chemotherapy use. Preliminary results suggest that fewer patients in the ctDNA-guided arm required adjuvant therapy with a lower risk of recurrence in patients with undetectable post-operative ctDNA [51].
The SYNCOPE study (NCT04842006) is a randomised controlled treatment trial aiming to reduce both over-treatment and metastatic disease progression in patients with rectal cancer. Participants with high-risk features will be randomised into two groups: early systemic chemotherapy followed by ctDNA and organoid-guided adjuvant therapy, or conventional treatment. Primary outcomes include RFS and ctDNA positivity rates in post-operative patients within the conventional treatment arm not exposed to chemotherapy.
The REVEAL study (NCT05674422) is a prospective multi-institutional study evaluating response to total neoadjuvant therapy (TNT) with liquid biopsy in eligible patients with LARC. The study aims to investigate the role of ctDNA in the prediction of relapse in this cohort of patients followed by a watch and wait programme or TME depending on the response to initial treatment.
Finally, CINTS-R (NCT05601505) is a multi-institutional randomised controlled trial aiming to evaluate outcomes of ctDNA-guided neoadjuvant treatment in patients with LARC. Treatment groups will receive either NCRT followed by immunotherapy, NCRT alone or TNT depending on mutation status following NGS of tumour tissue and variant allele frequency (VAF) of ctDNA. Control groups will receive standard NCRT only.
Discussion
Our systematic review of the current available literature demonstrates the significant potential of ctDNA as a prognostic biomarker in patients with LARC. Particularly, ctDNA detection post-neoadjuvant therapy or post-operatively was associated with an increased risk of recurrence, suggesting its utility in predicting disease progression and informing treatment decisions. Meta-analysis of available data further supported these findings, indicating a significantly higher hazard ratio for recurrence in patients with detectable ctDNA at these critical time points. These results align and extend upon existing literature on ctDNA in various cancer types, highlighting its potential as a non-invasive biomarker for monitoring disease burden and treatment response in a multitude of malignancies [4,5,6,7,8].
Despite its promise, ctDNA analysis is not without limitations [52]. Technical challenges, such as low concentrations of ctDNA in circulation and the need for sensitive detection methods, can impede accurate assessment. Similarly, tumour heterogeneity and clonal evolution may further complicate interpretation, potentially leading to false-negative or false-positive results [53]. Moreover, the lack of standardised protocols for ctDNA analysis and variability in assay performance across studies underscore the need for methodological standardisation and validation [54].
While our systematic review provides valuable insights into the role of ctDNA in LARC, several limitations warrant consideration. Firstly, studies included in our review demonstrated heterogeneity in terms of methodology, patient populations and outcome measures, which may have introduced bias and confounded interpretation. Additionally, the reliance on published data may have led to publication bias, with studies reporting significant findings more likely to be included. Despite these limitations, the findings of our review have significant implications for future research and clinical practice. The consistent association between ctDNA detection and adverse outcomes in LARC suggests its potential as a prognostic biomarker for risk stratification and treatment decision-making. Integrating ctDNA analysis into routine clinical practice could facilitate personalised treatment strategies, including the identification of high-risk patients who may benefit from intensified therapy or closer surveillance [55]. Furthermore, ongoing research efforts aimed at refining ctDNA-based assays and elucidating the underlying biological mechanisms driving ctDNA release and clearance are crucial for maximising its clinical utility [56].
Future research in this field should prioritise several key areas to address current knowledge gaps and optimise the implementation of ctDNA analysis in clinical practice. Firstly, large-scale prospective studies with standardised methodologies are needed to validate the prognostic utility of ctDNA across diverse patient populations and treatment settings [57]. Additionally, efforts to standardise ctDNA assays, including sample collection, processing and analysis protocols, are essential to ensure the reproducibility and comparability of results. Finally, collaborative efforts to establish international consortia and biobanks for ctDNA research could facilitate data sharing and accelerate progress towards clinical implementation [58].
Conclusion
Our systematic review provides evidence supporting the prognostic utility of ctDNA in patients with LARC, particularly in identifying patients at higher risk of disease recurrence in the post-neoadjuvant and post-operative periods.
Data availability
No datasets were generated or analysed during the current study.
References
Kokelaar RF, Evans MD, Davies M, Harris DA, Beynon J (2016) Locally advanced rectal cancer: management challenges. Onco Targets Ther 9:6265–6272
Smith JJ, Garcia-Aguilar J (2015) Advances and challenges in treatment of locally advanced rectal cancer. J Clin Oncol 33(16):1797–1808
Ryan EJ, Creavin B, Sheahan K (2020) Delivery of personalized care for locally advanced rectal cancer: incorporating pathological, molecular genetic, and immunological biomarkers into the multimodal paradigm. Front Oncol 10:1369
Chen C, Douglas MP, Ragavan MV, Phillips KA, Jansen JP (2023) Clinical validity and utility of circulating tumor DNA (ctDNA) testing in advanced non-small cell lung cancer (aNSCLC): a systematic literature review and meta-analysis. medRxiv [Preprint]. https://doi.org/10.1101/2023.10.27.23297657 (PMID: 37961510; PMCID: PMC10635208)
Guven DC, Sahin TK, Yildirim HC, Aktepe OH, Dizdar O, Yalcin S (2021) A systematic review and meta-analysis of the association between circulating tumor DNA (ctDNA) and prognosis in pancreatic cancer. Crit Rev Oncol Hematol 168:103528
Wullaert L, van Rees JM, Martens JWM, Verheul HMW, Grünhagen DJ, Wilting SM, Verhoef C (2023) Circulating tumour DNA as biomarker for colorectal liver metastases: a systematic review and meta-analysis. Cells 12(21):2520. https://doi.org/10.3390/cells12212520 (PMID: 37947598; PMCID: PMC10647834)
Sun X, Abrahamson P, Ballew N, Kalilani L, Phiri K, Bell KF et al (2023) The utility of ctDNA in lung cancer clinical research and practice: a systematic review and meta-analysis of clinical studies. Cancer Invest 41(6):571–592
Dizdarevic E, Hansen TF, Jakobsen A (2022) The prognostic importance of ctDNA in rectal cancer: a critical reappraisal. Cancers 14(9):2252. https://doi.org/10.3390/cancers14092252 (PMID: 35565381; PMCID: PMC9101261)
Pessoa LS, Heringer M, Ferrer VP (2020) ctDNA as a cancer biomarker: a broad overview. Crit Rev Oncol Hematol 155:103109
Gorgannezhad L, Umer M, Islam MN, Nguyen NT, Shiddiky MJA (2018) Circulating tumor DNA and liquid biopsy: opportunities, challenges, and recent advances in detection technologies. Lab Chip 18(8):1174–1196
Lebofsky R, Decraene C, Bernard V, Kamal M, Blin A, Leroy Q et al (2015) Circulating tumor DNA as a non-invasive substitute to metastasis biopsy for tumor genotyping and personalized medicine in a prospective trial across all tumor types. Mol Oncol 9(4):783–790
Postel M, Roosen A, Laurent-Puig P, Taly V, Wang-Renault SF (2018) Droplet-based digital PCR and next generation sequencing for monitoring circulating tumor DNA: a cancer diagnostic perspective. Expert Rev Mol Diagn 18(1):7–17
Satam H, Joshi K, Mangrolia U, Waghoo S, Zaidi G, Rawool S, Thakare RP, Banday S, Mishra AK, Das G, Malonia SK (2023) Next-generation sequencing technology: current trends and advancements. Biology 12(7):997. https://doi.org/10.3390/biology12070997 (Erratum in: Biology (Basel). 2024 Apr 24;13(5): PMID: 37508427; PMCID: PMC10376292)
Gezer U, Bronkhorst AJ, Holdenrieder S (2022) The clinical utility of droplet digital PCR for profiling circulating tumor DNA in breast cancer patients. Diagnostics 12(12):3042. https://doi.org/10.3390/diagnostics12123042 (PMID: 36553049; PMCID: PMC9776872)
Dymond JS (2013) Explanatory chapter: quantitative PCR. Methods Enzymol 529:279–289
Nakamura K (2016) Circulating Tumor DNA (ctDNA) Detection using BEAMing and its clinical significance. Rinsho Byori 64(4):400–6
Mouliere F, Chandrananda D, Piskorz AM, Moore EK, Morris J, Ahlborn LB, Mair R, Goranova T, Marass F, Heider K, Wan JCM, Supernat A, Hudecova I, Gounaris I, Ros S, Jimenez-Linan M, Garcia-Corbacho J, Patel K, Østrup O, Murphy S, Eldridge MD, Gale D, Stewart GD, Burge J, Cooper WN, van der Heijden MS, Massie CE, Watts C, Corrie P, Pacey S, Brindle KM, Baird RD, Mau-Sørensen M, Parkinson CA, Smith CG, Brenton JD, Rosenfeld N (2018) Enhanced detection of circulating tumor DNA by fragment size analysis. Sci Transl Med 10(466):eaat4921. https://doi.org/10.1126/scitranslmed.aat4921 (PMID: 30404863; PMCID: PMC6483061)
Khatami F, Tavangar SM (2018) Circulating tumor DNA (ctDNA) in the era of personalized cancer therapy. J Diabetes Metab Disord 17(1):19–30
Martín-Carnicero A, Ramalle-Gomara E, Rubio-Mediavilla S, Alonso-Lago M, Zorrilla-Larraga M, Manrique-Abós I, de las Heras-Dueña ME, Larrayoz IM, Martínez A (2022) Prognostic and predictive biomarkers in patients with Locally Advanced Rectal Cancer (LARC) treated with preoperative chemoradiotherapy. J Clin Med 11:6091. https://doi.org/10.3390/jcm11206091
Valenza C, Trapani D, Curigliano G (2022) Circulating tumour DNA dynamics for assessment of molecular residual disease and for intercepting resistance in breast cancer. Curr Opin Oncol 34(6):595–605
Page MJ, McKenzie JE, Bossuyt PM, Boutron I, Hoffmann TC, Mulrow CD et al (2021) The PRISMA 2020 statement: an updated guideline for reporting systematic reviews. BMJ 372:n71
Covidence systematic review software. Veritas Health Innovation, Melbourne, Australia
Stang A (2010) Critical evaluation of the Newcastle-Ottawa scale for the assessment of the quality of nonrandomized studies in meta-analyses. Eur J Epidemiol 25(9):603–605
Hozo SP, Djulbegovic B, Hozo I (2005) Estimating the mean and variance from the median, range, and the size of a sample. BMC Med Res Methodol 5:13
Agostini M, Pucciarelli S, Enzo MV, Del Bianco P, Briarava M, Bedin C et al (2011) Circulating cell-free DNA: a promising marker of pathologic tumor response in rectal cancer patients receiving preoperative chemoradiotherapy. Ann Surg Oncol 18(9):2461–2468
Alden SL, Lee V, Narang AK, Meyer J, Gearhart SL, Christenson ES (2024) Circulating tumor DNA to predict radiographic and pathologic response to total neoadjuvant therapy in locally advanced rectal cancer. Oncologist 29(3):e414–e418
Appelt AL, Andersen RF, Lindebjerg J, Jakobsen A (2020) Prognostic value of serum NPY hypermethylation in neoadjuvant chemoradiotherapy for rectal cancer: secondary analysis of a randomized trial. Am J Clin Oncol 43(1):9–13
Boysen AK, Wettergren Y, Sorensen BS, Taflin H, Gustavson B, Spindler KG (2017) Cell-free DNA levels and correlation to stage and outcome following treatment of locally advanced rectal cancer. Tumour Biol 39(11):1010428317730976
Guo ZW, Xiao WW, Yang XX, Yang X, Cai GX, Wang XJ et al (2020) Noninvasive prediction of response to cancer therapy using promoter profiling of circulating cell-free DNA. Clin Transl Med 10(5):e174
Hofste LSM, Geerlings MJ, von Rhein D, Rutten H, Westenberg AH, Weiss MM et al (2023) Circulating tumor DNA detection after neoadjuvant treatment and surgery predicts recurrence in patients with early-stage and locally advanced rectal cancer. Eur J Surg Oncol 49(7):1283–1290
Jiang XF, Zhang BM, Du FQ, Guo JN, Wang D, Li YE et al (2022) Exploring biomarkers for prognosis and neoadjuvant chemosensitivity in rectal cancer: multi-omics and ctDNA sequencing collaboration. Front Immunol 13:1013828
Khakoo S, Carter PD, Brown G, Valeri N, Picchia S, Bali MA et al (2020) MRI tumor regression grade and circulating tumor DNA as complementary tools to assess response and guide therapy adaptation in rectal cancer. Clin Cancer Res 26(1):183–192
Liu W, Li Y, Tang Y, Song Q, Wang J, Li N et al (2022) Response prediction and risk stratification of patients with rectal cancer after neoadjuvant therapy through an analysis of circulating tumour DNA. EBioMedicine 78:103945
McDuff SGR, Hardiman KM, Ulintz PJ, Parikh AR, Zheng H, Kim DW, Lennerz JK, Hazar-Rethinam M, Van Seventer EE, Fetter IJ, Nadres B, Eyler CE, Ryan DP, Weekes CD, Clark JW, Cusack JC, Goyal L, Zhu AX, Wo JY, Blaszkowsky LS, Allen J, Corcoran RB, Hong TS (2021) Circulating tumor DNA predicts pathologic and clinical outcomes following neoadjuvant chemoradiation and surgery for patients with locally advanced rectal cancer. JCO Precis Oncol 5:123–132. https://doi.org/10.1200/PO.20.00220 (PMID: 34250394; PMCID: PMC8232395)
Morais M, Fonseca T, Melo-Pinto D, Prieto I, Vilares AT, Duarte AL, Leitão P, Cirnes L, Machado JC, Carneiro S (2023) Evaluation of ctDNA in the prediction of response to neoadjuvant therapy and prognosis in locally advanced rectal cancer patients: a prospective study. Pharmaceuticals 16(3):427. https://doi.org/10.3390/ph16030427 (PMID: 36986526; PMCID: PMC10057108)
Murahashi S, Akiyoshi T, Sano T, Fukunaga Y, Noda T, Ueno M et al (2020) Serial circulating tumour DNA analysis for locally advanced rectal cancer treated with preoperative therapy: prediction of pathological response and postoperative recurrence. Br J Cancer 123(5):803–810
Pazdirek F, Minarik M, Benesova L, Halkova T, Belsanova B, Macek M et al (2020) Monitoring of early changes of circulating tumor DNA in the plasma of rectal cancer patients receiving neoadjuvant concomitant chemoradiotherapy: evaluation for prognosis and prediction of therapeutic response. Front Oncol 10:1028
Roesel R, Epistolio S, Molinari F, Saletti P, De Dosso S, Valli M et al (2022) A pilot, prospective, observational study to investigate the value of NGS in liquid biopsies to predict tumor response after neoadjuvant chemo-radiotherapy in patients with locally advanced rectal cancer: the LiBReCa study. Front Oncol 12:900945
Schou JV, Larsen FO, Sorensen BS, Abrantes R, Boysen AK, Johansen JS et al (2018) Circulating cell-free DNA as predictor of treatment failure after neoadjuvant chemo-radiotherapy before surgery in patients with locally advanced rectal cancer. Ann Oncol 29(3):610–615
Sclafani F, Chau I, Cunningham D, Hahne JC, Vlachogiannis G, Eltahir Z et al (2018) KRAS and BRAF mutations in circulating tumour DNA from locally advanced rectal cancer. Sci Rep 8(1):1445
Sun W, Sun Y, Zhu M, Wang Z, Zhang H, Xin Y et al (2014) The role of plasma cell-free DNA detection in predicting preoperative chemoradiotherapy response in rectal cancer patients. Oncol Rep 31(3):1466–1472
Tie J, Cohen JD, Wang Y, Li L, Christie M, Simons K et al (2019) Serial circulating tumour DNA analysis during multimodality treatment of locally advanced rectal cancer: a prospective biomarker study. Gut 68(4):663–671
Truelsen CG, Kronborg CS, Sorensen BS, Callesen LB, Spindler KG (2022) Circulating cell-free DNA as predictor of pathological complete response in locally advanced rectal cancer patients undergoing preoperative chemoradiotherapy. Clin Transl Radiat Oncol 36:9–15
Vidal J, Casadevall D, Bellosillo B, Pericay C, Garcia-Carbonero R, Losa F et al (2021) Clinical impact of presurgery circulating tumor DNA after total neoadjuvant treatment in locally advanced rectal cancer: a biomarker study from the GEMCAD 1402 trial. Clin Cancer Res 27(10):2890–2898
Wang Y, Yang L, Bao H, Fan X, Xia F, Wan J et al (2021) Utility of ctDNA in predicting response to neoadjuvant chemoradiotherapy and prognosis assessment in locally advanced rectal cancer: a prospective cohort study. PLoS Med 18(8):e1003741
Zhou J, Wang C, Lin G, Xiao Y, Jia W, Xiao G et al (2021) Serial circulating tumor DNA in predicting and monitoring the effect of neoadjuvant chemoradiotherapy in patients with rectal cancer: a prospective multicenter study. Clin Cancer Res 27(1):301–310
Chan HT, Nagayama S, Otaki M, Chin YM, Fukunaga Y, Ueno M et al (2022) Tumor-informed or tumor-agnostic circulating tumor DNA as a biomarker for risk of recurrence in resected colorectal cancer patients. Front Oncol 12:1055968
Parikh AR, Van Seventer EE, Siravegna G, Hartwig AV, Jaimovich A, He Y et al (2021) Minimal residual disease detection using a plasma-only circulating tumor DNA assay in patients with colorectal cancer. Clin Cancer Res 27(20):5586–5594
Gong J, Hendifar A, Gangi A, Zaghiyan K, Atkins K, Nasseri Y, Murrell Z, Figueiredo JC, Salvy S, Haile R, Hitchins M (2021) Clinical applications of minimal residual disease assessments by tumor-informed and tumor-uninformed circulating tumor DNA in colorectal cancer. Cancers 13(18):4547. https://doi.org/10.3390/cancers13184547 (PMID: 34572774; PMCID: PMC8471730)
Tie J, Wang Y, Tomasetti C, Li L, Springer S, Kinde I et al (2016) Circulating tumor DNA analysis detects minimal residual disease and predicts recurrence in patients with stage II colon cancer. Sci Transl Med 8(346):346ra92
Tie J, Cohen JD, Wang Y, Ginestet PG, Wong R, Shapiro JD et al (2024) Circulating tumor DNA analysis informing adjuvant chemotherapy in locally advanced rectal cancer: the randomized AGITG DYNAMIC-Rectal study. J Clin Oncol 42(3_suppl):12
Dang DK, Park BH (2022) Circulating tumor DNA: current challenges for clinical utility. J Clin Invest. 132(12):e154941. https://doi.org/10.1172/JCI154941 (PMID: 35703177; PMCID: PMC9197509)
Spoor J, Eyck BM, Atmodimedjo PN, Jansen M, Helmijr JCA, Martens JWM et al (2021) Liquid biopsy in esophageal cancer: a case report of false-positive circulating tumor DNA detection due to clonal hematopoiesis. Ann Transl Med 9(15):1264
Allan Z, Liu DS, Lee MM, Tie J, Clemons NJ (2024) A practical approach to interpreting circulating tumor DNA in the management of gastrointestinal cancers. Clin Chem 70(1):49–59
Wehrle J, Philipp U, Jolic M, Follo M, Hussung S, Waldeck S, Deuter M, Rassner M, Braune J, Rawluk J, Greil C, Waller CF, Becker H, Duque-Afonso J, Illert AL, Fritsch RM, Meiss F, Duyster J, von Bubnoff N, Scherer F (2020) Personalized treatment selection and disease monitoring using circulating tumor DNA profiling in real-world cancer patient management. Diagnostics 10(8):550. https://doi.org/10.3390/diagnostics10080550 (PMID: 32748806; PMCID: PMC7459590)
Sanchez-Herrero E, Serna-Blasco R, Robado de Lope L, Gonzalez-Rumayor V, Romero A, Provencio M (2022) Circulating tumor DNA as a cancer biomarker: an overview of biological features and factors that may impact on ctDNA analysis. Front Oncol 12:943253
Stadler JC, Belloum Y, Deitert B, Sementsov M, Heidrich I, Gebhardt C et al (2022) Current and future clinical applications of ctDNA in immuno-oncology. Cancer Res 82(3):349–358
Cree IA, Indave Ruiz BI, Zavadil J, McKay J, Olivier M, Kozlakidis Z et al (2021) The international collaboration for cancer classification and research. Int J Cancer 148(3):560–571
Acknowledgements
Supported by the Joly Leadership Fund.
Funding
Open Access funding provided by the IReL Consortium
Author information
Authors and Affiliations
Contributions
Conceptualisation: NOS, CG, JOS, GOK, BM, SOT, JL, DG, PMcC, MK; formal analysis: NOS, HCT, EK, KS, MON; investigation: NOS, HCT, EK, KS, MON; methodology: NOS, HCT, EK, KS, MON: supervision: CG, JOS, GOK, BM, SOT, JL, DG, PMcC, MK; visualisation: NOS, HCT, EK, KS, MK; writing—original draft: NOS, HCT, EK, KS, MON, CG, JOS, GOK, BM, SOT, JL, DG, PMcC, MK; writing—review and editing: NOS, HCT, EK, KS, MON, CG, JOS, GOK, BM, SOT, JL, DG, PMcC, MK.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Systematic review: ethical approval was obtained by individual studies to perform their research. All research was performed in accordance with the Declaration of Helsinki.
Consent for publication
N/A.
Competing interests
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors. The authors report no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
O’Sullivan, N.J., Temperley, H.C., Kyle, E.T. et al. Assessing circulating tumour DNA (ctDNA) as a prognostic biomarker in locally advanced rectal cancer: a systematic review and meta-analysis. Int J Colorectal Dis 39, 82 (2024). https://doi.org/10.1007/s00384-024-04656-1
Accepted:
Published:
DOI: https://doi.org/10.1007/s00384-024-04656-1