Abstract
Oomycetes are a ubiquitous protistan lineage including devastating crop parasites. Although their ecology in agrosystems has been widely studied, little is known of their distribution in natural and semi-natural ecosystems and how they respond to edaphic and environmental factors. We provide here a baseline of the diversity and distribution of soil oomycetes, classified by lifestyles (biotrophy, hemibiotrophy and saprotrophy), at the landscape scale in temperate grassland and forest. From 600 soil samples, we obtained 1148 operational taxonomy units representing ~ 20 million Illumina reads (region V4, 18S rRNA gene). We found a majority of hemibiotrophic plant pathogens, which are parasites spending part of their life cycle as saprotrophs after the death of the host. Overall both grassland and forest constitute an important reservoir of plant pathogens. Distance-based RDA models identified soil type and mineral soil C/N ratio as the most influential factors in shaping oomycete communities in grassland and forest. Edaphic conditions and human-induced management intensification in forest triggered opposite responses in the relative abundances of obligate biotrophs and hemibiotrophs, suggesting different ecological requirements of these two lifestyles.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Oomycetes are protists (phylum Stramenopiles or Heterokonta) ubiquitous and widespread in terrestrial (Geisen et al. 2015; Lara and Belbahri 2011; Singer et al. 2016), freshwater (Duffy et al. 2015) and marine ecosystems (Garvetto et al. 2018). In terrestrial ecosystems, oomycetes occur as pathogens of plants and other eukaryotes and, less commonly, as saprotrophs (Marano et al. 2016), with plant pathogens representing more than 60% of the oomycete taxa (Thines and Kamoun 2010). Well-known examples are the soil-borne downy mildews with genera like Phytophthora and Pythium and the white rusts (Albugo) on plant leaves (Savory et al. 2015). The genus Pythium is one of the most important soil-borne plant pathogens, being ubiquitous and with an extremely wide host range, attacking the roots of thousands of different plant species (Beakes and Thines 2016). Phytophthora, the “plant destroyer”, is responsible for the widespread rapid tree decline (Hayden et al. 2013) and for damages to important crops like soybean, tomato, grapevine and tobacco (Lebeda et al. 2008). Because of their negative economic impact, oomycetes are well-studied in silico and as many as 67 species have their genome available in public databases (https://www.ncbi.nlm.nih.gov/genome, last accessed 28 March 2020). Although they affect forest ecosystems worldwide (Packer and Clay 2000) and are also common pathogens in grasslands (Foley and Deacon 1985), their occurrence in natural habitats and their ecological role in maintaining plant species diversity (Bever et al. 2015) is still poorly explored compared to agroecosystems.
Oomycota is not a functionally homogeneous group, since it includes lifestyles as diverse as saprotrophy, biotrophy and hemibiotrophy—the two latter being two forms of parasitism. Free-living saprotrophs live on organic matter or dead tissues. Lifestyles of pathogens include obligate biotrophy which requires living plant tissue (e.g. Lagena, Peronospora and Plasmopara) and hemibiotrophy (e.g. Phytophthora and Pythium) characterised by an initial biotrophic infection, followed by necrotrophy on killed host tissue (Ah-Fong et al. 2019; Pandaranayaka et al. 2019). It is important to note that these categories describe life phases rather than the microorganisms themselves (Lorang 2019). Many hemibiotrophic intermediate states between biotrophy and necrotrophy are recognised (Spanu and Panstruga 2017), varying in the length of the latent period, the degree of inflicted damage and the susceptibility of the host (Précigout et al. 2020). In addition, most species of hemibiotrophs can live as saprotrophs in the soil, even in the absence of the host plant (Lifshitz and Hancock 1983), and recently, species of Pythium have been shown to play an important role in litter degradation (Kramer et al. 2016). We did not expect that such fundamentally different lifestyles would unequivocally respond to environmental shaping forces, nor displaying homogeneous biogeographies: whenever functional traits are considered, protistan taxa show differential distributions accordingly (Faure et al. 2019; Fiore-Donno et al. 2019, 2020; Jassey et al. 2016). We hypothesised that hemibiotrophs would be more abundant in the habitats studied than the more specialised obligate biotrophs. In addition, we expected that soils rich in organic carbon would harbour less plant parasites (Hayden et al. 2013), while fertilisation would increase them (Löbmann et al. 2016).
Recent large-scale studies, using next-generation sequencing, have revealed that Oomycota represented c. 5–10% of the protistan metatranscriptomics in forest and grassland soils and to a lesser extent in peatlands (Geisen et al. 2015). A study conducted only on peat bogs, using ad hoc primers, revealed nevertheless the occurrence of 34 phylotypes, most of which could not be assigned to known species (Singer et al. 2016). Few plant-associated oomycetes were found in the roots of oaks (Sapp et al. 2019), in that of Arabidopsis thaliana (Sapp et al. 2018) and in various other plant roots along a glacier chronosequence (Dickie et al. 2019). Their complex life cycle, including a stage with large, mycelium-like structures protected by cell walls, the occurrence of motile cells for dispersion and thick-walled resting stages (Beakes and Thines 2016), may explain their adaptability and dispersal potential. This suggests that a wider diversity of Oomycetes should be detectable by large-scale metabarcoding (taxon identification using a short fragment of a gene from environmental DNAs) studies in soil than the currently known c. 2000 species, of which 95% belong to the two crown groups, the Saprolegniales and Peronosporales (Thines and Choi 2016).
Our aim was to assess: (i) the biodiversity of oomycetes in a large-scale environmental survey in grasslands and forests of the Biodiversity Exploratories, Germany; (ii) the influence of edaphic, environmental and anthropogenic factors on the composition of oomycete communities classified according to their lifestyle (obligate biotrophy, hemibiotrophy or saprotrophy) and substrate preference (animal- or plant-associated). Providing such a detailed baseline is an important contribution to assess the roles of plant pathogens for ecosystem functioning.
Materials and methods
Study site, soil sampling and DNA extraction
Our study took place in three German Biodiversity Exploratories, i.e. the Biosphere Reserve Schorfheide-Chorin in the State of Brandenburg, the National Park Hainich and its surroundings in the State of Thuringia and the Biosphere Reserve Schwäbische Alb in the State of Baden-Württemberg (Fischer et al. 2010). Each exploratory comprises 50 grassland sites from extensive pastures to highly fertilised meadows and 50 differently managed forest sites, composed mainly of beech (Fagus sylvatica) sometimes mixed with spruce (Picea abies) (detailed information, maps and photos at https://www.bexis.uni-jena.de/PublicData/About.aspx, last accessed July 2020). Each site contains a study plot of 20 × 20 m. From all study plots, 300 soil samples were collected in a coordinated joint sampling campaign within 14 days in April 2011 and a second one in April 2017. From each plot, 14 soil cores of 5 cm diameter were taken every 3 m along two transects of 20 m (grassland) and 40 m (forest) each, oriented north-south and east-west, employing a soil corer. The surface layer (0–10 cm) was collected, after removing plants, pebbles and conspicuous roots. Soil cores from each plot were sieved (2-mm mesh size), mixed, homogenised and immediately frozen for further analysis of edaphic properties. Soil DNA was extracted from 400 mg of soil (in 2011 and 2017), 3 to 6 times, using the DNeasy PowerSoil Kit (Qiagen GmbH, Hilden, Germany) following the manufacturer’s protocol, to obtain a sufficient amount to be shared between research groups of the Biodiversity Exploratories. Edaphic properties were determined by the Biodiversity Exploratories core project and are available at https://www.bexis.uni-jena.de/PublicData/About.aspx (last accessed July 2020). Briefly, they were determined as follows: pH with a soil:solution (0.01 M CaCl2) ratio of 1:2.5; total C and N concentrations using an elemental analyser; inorganic C was determined with the same elemental analyser after removal of organic carbon by ignition at 450 °C for 16 h; soil texture by the percentage of sand (particles of 2–0.063 mm), silt (0.063–0.002 mm) and clay (< 0.002 mm) by sieving and sedimentation, after removal of soil organic matter with hydrogen peroxide.
Primer and barcode design—amplification
We designed primers to target the ITS1, because it better allows to distinguish between species (although not in the closely related species of some groups of the polyphyletic Pythium) (Steciow et al. 2014), and its reference database is rich (c. 600 species). Potential problems with the ITS1 marker are the variation in length (216–534 bp, the longest sequences in Plasmopara). Because Oomycota is a large and diverse assemblage, we could design primers to match the two major clades (in number of species and ecological importance), the Saprolegniales and the Peronosporales, missing the small basal lineages, i.e. the marine Eurychasmales, Haliphthorales and Olpidiopsidales and partially the nematode infecting Haptoglossales. We discarded other potential markers such as the V4 region of the small subunit of the ribosomal RNA gene because it does not display enough intraspecific variation, and the mitochondrial cytochrome oxidase COX2 because it has a very high AT-rich content, which could negatively interfere with the quality of the Illumina run.
Primers were designed using an alignment of 2941 Oomycetes ITS1 sequences. To build it, we downloaded all ITS sequences > 500 bp from GenBank RefSeq (accessed 24.08.2017), clustered them at 96% similarity using cd-hit 454 (Beifang et al. 2010). This database was then used for Blast purposes and is provided (Supplementary information S1). For primer design, we removed sequences with ambiguities, aligned them with MAFFT (Katoh and Standley 2013); the alignment was refined by hand using Bioedit (Hall 1999) and cut to the fragment of interest. Our forward primers were located at the very end of the SSU (a bit downstream the widely used ITS6) (Foster et al. 2020; Sapkota and Nicolaisen 2015) and similar to several primers overlapping the beginning of the ITS1, e.g. ITS1-ofw (Liebe et al. 2016), ITS1oo (Riit et al. 2016) and OOMUP18Sc (Lievens et al. 2004). The reverse primer in the 5.8S gene was partially overlapping the primers ITS1-orw (Liebe et al. 2016) and ITS-3oo (Riit et al. 2016). The widely used reverse primer ITS4 (Foster et al. 2020; Lievens et al. 2004; Riit et al. 2016; Sapkota and Nicolaisen 2015) is located in the LSU after the ITS2, and thus not suitable for targeting only the ITS1; in addition, the amplified fragment is too long for Illumina sequencing. We designed three primers (two forward primers partially overlapping) to be used in two successive semi-nested PCRs. The primers did not form any hairpin, primer-dimers with themselves or with each other, and their hybridisation temperatures were similar. The length of the targeted fragment varied from 206 to 534 bp. The first PCR was conducted with the primers S1777F (non-specific - 5′ GGTGAACCTGCGGAAGGA 3′, located at the end of the 18S gene) (1777 is the starting position in the SSU sequence Saccharomyces cerevisiae Z75578) and 58SOomR (oomycete-specific - 5′ TCTTCATCGDTGTGCGAGC 3′). The second PCR was conducted with barcoded primers S1786StraF (specific for Stramenopiles - 5′ GCGGAAGGATCATTACCAC 3′) and the 58SOomR as before. The barcodes consisted of eight-nucleotide-long sequences appended to the 5′-ends of both the forward and the reverse primers, because tagging only one primer leads to extensive mistagging (Esling et al. 2015). To design the barcodes, we first used barcrawl (Frank 2009) to obtain a list of barcodes with a balanced nucleotide content (no homopolymers), not folding on themselves and to themselves and the attached primer (no “hairpin”), not forming heteroduplexes with the corresponding primer and having at least 3 bases differences between them. In addition, using custom R scripts, we selected from the previous list the barcodes that did not match the consensus of the reference alignment flanking the primer region (forward: GTGAACCT, reverse: VCGCTGCG) and without cross-dimerisation between each combination of primer+barcodes. We designed 18 barcoded versions for the forward and the reverse primers allowing for 324 possible combinations to label samples of which only 150 were used, since it is advisable to leave a proportion of unused combinations to decrease mistagging (Esling et al. 2015). Barcoded primers were specifically ordered for NGS application to Microsynth (Wolfurt, Austria) (Table S1).
For the amplification, we incorporated 1 μl of 1:10 soil DNA template for the first PCR and 1 μl of the resulting amplicon as a template for a following semi-nested PCR. We employed the following final concentrations: GreenTaq polymerase (Fermentas, Canada) 0.01 units, buffer 1×, dNTPs 0.2 mM and primers 1 μM. The thermal programme consisted of an initial denaturation step at 95 °C for 2 min, 24 cycles at 95 °C for 30 s, 52 °C for 30 s, 72 °C for 30 s; and a final elongation step at 72 °C for 5 min. The number of PCR cycles was kept at 24 since chimera formation arises dramatically after 25 cycles (Michu et al. 2010). All PCRs were conducted twice to reduce the possible artificial dominance of few amplicons by PCR competition (2 × 10 μl for the first and 2 × 27 μl for the second PCR), and the two amplicons were pooled after the second PCR.
Library preparation and sequencing
The amplicons were checked by electrophoresis and 25 μl of each were purified and normalised using SequalPrep Normalization Plate Kit (Invitrogen GmbH, Karlsruhe, Germany) to obtain a concentration of 1–2 ng/μl per sample, which were then pooled, totalling four libraries (forest 2011 and 2017 and grassland 2011 and 2017). During the library preparation, amplicons were end-repaired, small fragments were removed, 3′-ends were adenylated and Illumina adapters and sequencing primers were ligated (TruSeqDNA PCR-Free, Illumina Inc., San Diego, CA, USA). The library was quantified by qPCR, performed following the manufacturer’s instructions (KAPA SYBR® FAST qPCR Kit, Kapabiosystems, Wilmington, MA, USA) on a CFX96 Real Time System (Bio-Rad, Hercules, CA, USA). Sequencing was performed with a MiSeq v3 Reagent kit of 600 cycles on a MiSeq Desktop Sequencer (Illumina Inc., San Diego, CA, USA) at the University of Geneva (Switzerland), Department of Genetics and Evolution. Four runs were conducted in total.
Sequence processing
In each run, paired reads were assembled using mothur v.3.7 (Schloss et al. 2009) (which was also used in the following steps) allowing no difference in the primers and barcodes, no ambiguities and removing assembled sequences < 200 bp, with an overlap < 100 bp or with any mismatch in the overlap (Table 1). The quality check and removal/cutting of low-quality reads was conducted with the default parameters. Reads were sorted into samples via detection of the barcodes (Table S1), renamed and then the four runs were assembled. Sequences were clustered using vsearch v.1 (Rognes et al. 2016) in mothur, with abundance-based greedy clustering (agc) and a similarity threshold of 97% (Table 1). Rare OTUs (< 0.001% of the total sequences, in this case < 368 reads) were deleted, since we observed that they mostly represented artifactual reads (Fiore-Donno et al. 2018).
Using BLAST+ (Camacho et al. 2008) with an e value of 1e−1 and keeping only the best hit, sequences were identified on a custom ITS database (Supplementary information S1) and non-oomycetes sequences were removed (Table 1). Since the ITS sequences were too variable to be aligned, we performed the chimera check using UCHIME (Edgar et al. 2011) as implemented in mothur by taking into account the abundance of the OTUs and the OTUs identified as chimeric were removed. The results are shown as a table with the OTU abundance/site, their taxonomic assignment according to the best hit by Blast and their functional assignment (Table S2). The relative abundance of each oomycete taxonomic level was illustrated using Sankey diagram generator V1.2 (http://sankey-diagram-generator.acquireprocure.com/, last accessed Jan. 2020) and refined with the open-source vector graphic editor Inkscape (https://inkscape.org/en/, last accessed Jan. 2020).
Functional guilds
We compiled a table with functional traits of ecological importance, i.e. lifestyle and substrate. Oomycete lifestyles that are traditionally distinguished are saprotrophs—living on dead organic matter (e.g. decaying plants, insect exuviae), biotrophs (obligate parasites or pathogens) and hemibiotrophs (facultative parasites, switching from a parasitic to a saprotrophic lifestyle). We also collected information about the substrate—plant or animal—for saprotrophs as well as for pathogens (Table S2). We could only attribute functional traits to the level of the genus, since we clustered the OTUs at 97% similarity, and because the uncertainty of the attribution in the database increases at the species level. We attributed traits by searching in the relevant literature, providing all consulted references (Table S3). We could not find sufficient information about the dispersion mode—aerial, or solely through water and soil.
Statistical analyses
All statistical analyses were carried out within the R environment (R v. 3.5.1) (R Development Core Team 2014), on the OTU abundance/site table (Table S2), and the environmental parameters (Table S4 and Table S5), the latter normalised by the K-nearest neighbours. Unless otherwise specified, community analyses were performed with the package vegan (Oksanen et al. 2013). Alpha diversity: To evaluate if more sampling and sequencing effort would have revealed more OTU richness, we carried out an analysis based on OTU accumulation curves, function specaccum, method rarefaction and 1000 random permutations; species richness was extrapolated using the function specpool. Alpha diversity estimates were based on the relative abundances of OTUs (function decostand, method total); Shannon diversity and Pielou’s evenness were obtained with the function diversity. Beta diversity: Variation partitioning (function varpart applied to the Hellinger-transformed OTU dataset and using RDA, function rda) was performed to assess the proportion of explained beta diversity by the factors region, year of collection (2011, 2017) or ecosystem (grassland vs forest). Beta diversity between regions and ecosystems was inferred by principal coordinate analysis (PCoA, function cmdscale), using Bray-Curtis dissimilarities (function vegdist, method “bray”) on the relative abundances of OTUs, then plotted with the package ggplot2. A distance-based redundancy analysis (dbRDA) on Bray-Curtis dissimilarities (function dbrda) was used to investigate the effect of environmental factors on the beta diversity of oomycete communities. Parsimonious models were selected by the function ordistep with default parameters based on 999 permutations, and only significant results were shown in dbRDA. All available parameters were tested for co-correlation according to variance inflation factors (function vif.cca). The effects of environmental parameters on the relative abundances of the lifestyles were tested with general linear models (function glm, core package). They were then subjected to the general linear hypothesis test (function glht, package multcomp) with Tukey’s test for multiple comparisons of means and a heteroskedasticity-consistent covariance matrix estimation (function vcovHC, package sandwich).
Results
Sequencing
We obtained more than 22 million reads per run. During the sequencing, the overall quality was good, between 84 and 92% ≥ Q30. The reads had on average a length of c. 240 bp, and the overlap during assemblage was of c. 200 bp. In total, we obtained 1148 oomycete OTUs representing 20,501,201 sequences (Table 1) in 600 grasslands and forest sites, with on average 34,168 reads/site (minimum 460, maximum 108,910). Our primers also amplified c. 28% of sequences that could not be identified in the database. Because of the high variability of the ITS 1 region, no satisfactory automated alignment could be obtained, and the chimera check had to be performed with non-aligned sequences, a somewhat less stringent method, probably leaving a proportion of undetected chimeras in our database, although c. 7% were deleted (Table 1). The 1148 OTUs represented 319 unique Blast best hits. Only 31% of the OTUs were 97–100% similar to any known sequence (Fig. S1). The 30 most abundant OTUs (> 10,000 sequences) accounted for 73% of the total sequences, while 526 OTUs of < 1000 sequences contributed only to 1.6% of all sequences (Table S2). A database with the OTU abundance per sample, taxonomic assignment and estimated functional traits is provided (Table S2 and Table S3). Our sequencing and sampling efforts were sufficient, since the actual richness was reached after only 310,000 sequences (Fig. S2A) and at 200 samples (Fig. S2B), so that the observed distribution patterns would not have been influenced by undersampling.
Diversity of oomycetes
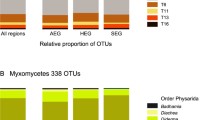
At a high taxonomic level, the majority of the OTUs could be assigned to the Peronosporales (73%) and only 21% to the Saprolegniales, with Leptomitales (5%) and Haptoglossales (1%) only marginally present, the latter probably because our primers did not match all of them (Fig. 1). We did not use the family rank because the traditional classification is not supported by most phylogenies and varies between authors. Among the 32 identified genera, the most common was by far Pythium (50%), followed by Saprolegnia (10%), Aphanomyces (6%), Phytophthora (5%) and Apodachlya (5%). The Haptoglossales (1%) contain only a dozen species, all of which are parasites of rotifers and bacterivorous nematodes (Beakes and Thines 2016). Representatives of the order Rhipidiales (largely saprotrophic and aquatic) are missing from the reference database (Supplementary information S1) and were thus not being found in our samples.
Sankey diagram showing the relative contribution of the OTUs to the taxonomic diversity. Taxonomical assignment is based on the best hit by BLAST. From left to right, names refer to phyla, orders (ending -ida) and genera. Numbers are percentages of OTUs abundance—OTUs representing < 1% are not shown
Functional guilds
A majority of hemibiotrophs was found (75–76%, 860 OTUs). The most abundant genera were pathogens of plants, i.e. Pythium (67% of the hemibiotrophs) and Phytophthora (6%). Abundant genera of animal pathogens were Saprolegnia (14%), Lagenidium (5%), Myzocytiopsis (3%) and Atkinsiella (2%) (Fig. 1, Fig. 2 and Table S2). The obligate biotrophs (10–11%) were less represented and were mostly parasites of plants. For example, Lagena (34% of the obligate biotrophs) is a root-infecting parasite of grasses, including cereals (Blackwell 2011), Peronospora (28%) is a downy mildew which parasitises a wide range of flowering plants, Plasmopara (14%) has a wide host spectrum in eudicots (Thines and Choi 2016) and Hyaloperonospora (9%) infects Brassicaceae. The saprotrophs, e.g. Apodachlya (70% of the saprotrophs) and Leptolegnia (14%), counted only for 6–8% of all OTUs. The relative proportions of each lifestyle were quite similar between forest and grassland (Fig. 2). Most oomycetes depended on plants (65%), more in grassland (69%) than in forest (66%). Animal-dependent oomycetes (21% in total) were slightly more abundant in forest (21%) than in grassland (17%) (Fig. 2).
Alpha and beta diversity
A total of 1007 and 1072 oomycete OTUs were retrieved in grassland and forest sites, respectively. The great majority of the OTUs (931, 81%) were shared between ecosystems, with only 76 unique to grassland and 141 to forest. Most OTUs were shared between regions, only 37 were missing in Schorfheide, five in Hainich and eight in Alb (Table S2). Alpha diversity, as revealed by Shannon indices, was significantly higher in grassland than in forest, while at the regional level, only Schorfheide had a significantly lower alpha diversity. Evenness of oomycete OTUs was higher in grassland than in forest, and higher in Alb and Hainich than in Schorfheide (Fig. S3). Despite the almost ubiquitous presence of oomycete OTUs in grasslands, PCoA revealed differences in beta diversity between communities, which can be explained by differences in the relative abundance of taxa. Both PCoA components, explaining 34% and 13% of the variance in Bray-Curtis distances, showed gradients from grassland to forest, although with some overlap (Fig. 3). The grassland communities were more similar between themselves than the forest communities, where the effect of the region was more pronounced, with Schorfheide standing out, especially its forest communities (Fig. 3).
Major drivers structuring oomycete communities
Variation partitioning among the three predictors indicated that ecosystem (grassland vs forest), region (Alb, Hainich and Schorfheide) and year of sampling (2011 and 2017) together accounted for 28.6% (adjusted R2) of the total variation in oomycete beta diversity. Ecosystem explained 16.3% of the variation, region 10.0%, and the year of sampling only 2.2%. Among the soil parameters which were measured in the framework of the Biodiversity Exploratories (Table S4 and Table S5), carbon content (total, organic and inorganic) and nitrogen (total N) were all co-correlated; they were tested separately, and based on a slightly higher significance in the models, organic carbon was kept. Among the three co-correlated soil components, sand was preferred over clay or silt, because of its recognised importance for oomycete occurrence. The soil and ecological parameters included in our analyses could explain more variance in forests (R2 10.5–25.3) than in grasslands (R2 2.6–5.9) (Table S6).
Edaphic properties: The most parsimonious models identified an influence of soil type in grassland as well as in forest (Table S6). In addition, in grassland, the soil C/N ratio and the organic C content were influential, but only for specific lifestyles or regions. In addition to soil type, soil pH and sand content were identified as explanatory factors in forest. Anthropogenic influences in grassland were the land use intensity (LUI) index and the mowing intensity. In forest, the main tree species and the forest management intensity were selected by the models. Other factors, selected by a minority of the models in forest, were forest developmental stage, organic C content, and C/N ratio (Table S6).
We further investigated the effects of these selected environmental parameters on each of the oomycete lifestyles, by comparing their relative abundances in the two ecosystems. Hemibiotrophs and biotrophs (and to a lesser extent, also saprotrophs) showed opposite responses to environmental parameters, both in grassland (Fig. S4) and forest (Fig. S5). Less significant differences were found in grassland than in forest, since the OTUs in the former communities were more evenly distributed (Fig. 3). The oomycete communities in grasslands of Schorfheide stood out with respect to Hainich, with slightly decreasing biotrophs and increasing saprotrophs (Fig. S4A), a tendency also partially reflected by the communities of specific soil types that were either only present in Hainich or in Schorfheide (Fig. S4B). Hemibiotrophs decreased at a higher LUI index (Fig. S4C). An increasing soil C/N ratio led to a relative decrease of hemibiotrophs and an increase of biotrophs (Fig. S4D). In soils with high organic carbon content, biotrophs decreased and saprotrophs increased (Fig. S4E).
In forest, hemibiotrophs and biotrophs together with saprotrophs showed opposite shifts in relative abundances patterns. At the regional scale, Schorfheide differed from Hainich and Alb by a decrease of hemibiotrophs and an increase of biotrophs and saprotrophs (Fig. S5A), a trend mirrored by the soil types unique to each region (Fig. S5B) and further by the tree species unique to Schorfheide, i.e. pine and oak (Fig. S5C). Hemibiotrophs were reduced, while biotrophs and saprotrophs slightly increased with increasing forest management intensity and increasing sand content (Fig. S5D and E). With an increasing soil C/N ratio in forests, hemibiotrophs decreased and biotrophs increased (as in grasslands) but also saprotrophs increased (Fig. S5F). Hemibiotrophs decreased in more acidic soils (mostly present in Schorfheide), while biotrophs and saprotrophs increased (Fig. S5G).
Discussion
Our data stemmed from a thorough sampling across three regions spanning a south-north gradient in Germany using taxon-specific primers. Based on the high sequencing depth (saturation was reached) (Fig. S2), we could detect detailed responses of each lifestyle to the ecological and edaphic factors involved in shaping oomycete distribution. The high percentage of OTUs not closely matching any published sequence (Fig. S1) suggests a significant hidden species richness not yet taxonomically recorded or sequenced in the ITS1 database. Consistently with previous studies on protists, a high alpha diversity and a low beta diversity were found (Fiore-Donno et al. 2019, 2020; Lentendu et al. 2018): almost all OTUs were shared between ecosystems and regions. This implies that community assembly of oomycetes is not limited by dispersal over countrywide distances. As a corollary, the remarkable differences in beta diversity are probably explained by contrasted responses of lifestyles (assigned to taxa) to environmental selection (Fiore-Donno et al. 2019).
Plant-associated hemibiotrophs are abundant in natural and semi-natural ecosystems: differences between grassland and forest and significance for agriculture
We found that plant-associated hemibiotrophic oomycetes, in particular Pythium and Phytophthora, the two most infamous destructive oomycete plant pathogens, dominated in natural and semi-natural ecosystems, which thus constitute a reservoir of plant pathogens with the potential to spread to neighbouring agroecosystems. The high abundance of the genus Pythium (Fig. 1), is in line with previous oomycete community studies (Riit et al. 2016; Sapkota and Nicolaisen 2015; Venter et al. 2017), and with Peronosporales being by far the largest order in Oomycota, comprising more than 1000 species (Thines 2014). Hemibiotrophs most probably benefit from their ability to alternate between saprotrophy and parasitism, while obligate biotrophs (e.g. Peronospora, Plasmopara), being more host-dependent, were less abundant (Fig. 2). Our study, in assessing the biogeography of oomycete pathogens across Germany, also contributes to a better understanding of the mechanisms of dispersal from natural reservoirs to agrosystems, and perhaps could contribute to better reveal the evolutionary processes leading to the emergence of new pathogens.
Grassland soils hosted more diverse and more evenly distributed oomycete communities than forest soils (Fig. S3). This is in accordance with a study comparing ecotypes across Wales, where the relative abundance of oomycetes decreased from crops and grasslands to forests and bogs (George et al. 2019). In this study and ours, Oomycetes do not follow the general pattern of soil microbes, that is a higher richness in forest compared to grassland, as it has been reported for bacteria and fungi in the Biodiversity Exploratories (Birkhofer et al. 2012; Foesel et al. 2014; Kaiser et al. 2016; Nacke et al. 2001) and for protists (Ferreira de Araujo et al. 2018; Zhao et al. 2018). In particular, in the same sites of the Biodiversity Exploratories, Cercozoa and Endomyxa were more diverse and more evenly distributed in forest than in grassland, except for the endomyxan plant parasites, which were absent from forest (Fiore-Donno et al. 2020). This suggests that temperate grasslands may be a more favourable environment for parasitic protists.
Obligate biotrophs and hemibiotrophs showed opposite responses to environmental and edaphic factors in forest
In forests, hemibiotrophs and obligate biotrophs showed opposite responses to a number of environmental factors, suggesting different ecological requirements of both functional groups. Nitrogen-poor, sandy and acidic soils, planted with pines and oaks, which are the type of soils common in Schorfheide, favoured biotrophs and saprotrophs and decreased hemibiotrophs (Fig. S5). The same combination of edaphic and ecological factors which led to a reduced abundance of hemibiotrophs favoured biotrophs and saprotrophs. This was a surprising result: we expected hemibiotrophs to show intermediate responses, while obligate parasites and saprotrophs would each represent opposite extremes of the response gradient. This assumption was based on the evolutionary distance—saprotrophy being the ancestral stage and biotrophy the last to evolve—and the substrate preferences. Our data clearly show that environmental factors differentially influence hemibiotrophs, obligate biotrophs and saprotrophs.
Soil texture is well known to influence the abundance and disease development of oomycetes. Waterlogging conditions that have been shown to strongly benefit oomycete pathogens (Gómez-Aparicio et al. 2012) occur less in sandy than in clayey soils. Accordingly, we found that hemibiotrophs strongly decreased in forest soils with a high sand content (Figs. 4 and S5E) and were more abundant in soils periodically inundated (Stagnosols) (Fig. S5B). Soils with high organic content, or soils amended with compost, have long been known to suppress a number of Pythium and Phytophthora species (Hayden et al. 2013), but the reason is not precisely known. One hypothesis is that carbon-rich soils harbour a wide diversity of microbes that may outcompete oomycetes, especially hemibiotrophic species with saprotrophic capacities, like some Pythium (Hayden et al. 2013). Not only carbon but also nitrogen content has an effect on soil suppressiveness, which occurs less frequently under high-nutrient conditions (Löbmann et al. 2016). Accordingly, relative abundances of hemibiotrophs, both in grassland and forest, decreased in nitrogen-poor soils with a high carbon content (but not that of biotrophs) (Figs. 4, S4D and S5F), while saprotrophs, being carbon-dependent, increased with C/N ratio in forest (Fig. S5F) and with organic C content in grassland (Fig. S4E). Thus, in order to successfully enhance soil suppressiveness, it is necessary to understand how particular management practices will differentially influence each key component of biodiversity (Löbmann et al. 2016; Schlatter et al. 2017). Here, we show that a more intensive forest management, as recorded in the Biodiversity Exploratories (including estimation of harvested trunks, invasive tree species and cut dead wood) will not increase the abundance of hemibiotrophs (Fig. 4).
Schematic illustration showing the positive or negative influences of the most influential ecological and edaphic parameters in forests (selected by the models in Table S5) and the relative abundances of the oomycetes lifestyles. For numeric parameters, minimum, average and maximum values are given. Forest developmental stage was estimated by the mean of the diameter of 100 trunks of well-developed trees of the dominant species
Concluding remarks
Studies on the ecology of microbial eukaryotes suffer from a lack of systematic basic information on the community composition and the factors shaping it which hampers predicting the effects of anthropogenic environmental changes. Providing a detailed baseline data on the occurrence of oomycete taxa, the ecology and the distribution of their lifestyles is an important contribution to the understanding of ecological processes and ecosystem functioning, a prerequisite for subsequent analyses linking them to the distribution and diversity of potential plant hosts.
Data availability
Raw sequences have been deposited in Sequence Read Archive (NCBI) SRA 10697395-8, Bioproject PRJNA513166 and the 1148 OTUs (representative sequences) under GenBank accession nos. MN268786 - MN269933.
References
Ah-Fong AMV, Kagda MS, Abrahamian M, Judelson HS (2019) Niche-specific metabolic adaptation in biotrophic and necrotrophic oomycetes is manifested in differential use of nutrients, variation in gene content, and enzyme evolution. PLoS Path 15:e1007729. https://doi.org/10.1371/journal.ppat.1007729
Beakes G, Thines M (2016) Hyphochytriomycota and Oomycota. In: Archibald JM, Simpson AGB, Slamovits CH, Margulis L, Melkonian M, Chapman CP, Corliss JO (eds) Handbook of the protists. Springer International Publishing AG, Basel, pp 435–505. https://doi.org/10.1007/978-3-319-32669-6_26-1
Beifang N, Limin F, Shulei S, Weizhong L (2010) Artificial and natural duplicates in pyrosequencing reads of metagenomic data. BMC Bioinf 11:187. https://doi.org/10.1186/1471-2105-11-187
Bever JD, Mangan SA, Alexander HM (2015) Maintenance of plant species diversity by pathogens. Annu Rev Ecol Syst 46:305–325. https://doi.org/10.1146/annurev-ecolsys-112414-054306
Birkhofer K, Schöning I, Alt F, Herold N, Klarner B, Maraun M, Marhan S, Oelmann Y, Wubet T, Yurkov A, Begerow D, Berne D, Buscot F, Daniel R, Diekötter T, Ehnes RB, Erdmann G, Fischer C, Foesel BU, Groh J, Gutknecht J, Kandeler E, Lang C, Lohaus G, Meyer A, Nacke H, Näther A, Overmann J, Polle A, Pollierer MM, Scheu S, Schloter M, Schulze E-D, Schulze W, Weinert J, Weisser WW, Wolters V, Schrumpf M (2012) General relationships between abiotic soil properties and soil biota across spatial scales and different land-use types. PLoS ONE 7:e43292. https://doi.org/10.1371/journal.pone.0043292
Blackwell WH (2011) The genus Lagena (Stramenopila: Oomycota), taxonomic history and nomenclature. Phytologia 93:157–167
Camacho C, Coulouris G, Avagyan V, Ma N, Papadopoulos J, Bealer K, Madden TL (2008) BLAST+: architecture and applications. BMC Bioinf 10:421. https://doi.org/10.1186/1471-2105-10-421
Dickie IA, Wakelin AM, Martínez-García LB, Richardson SJ, Makiola A, Tylianakis JM (2019) Oomycetes along a 120,000 year temperate rainforest ecosystem development chronosequence. Fungal Ecol 39:192–200. https://doi.org/10.1016/j.funeco.2019.02.007
Duffy MA, James TY, Longworth A (2015) Ecology, virulence, and phylogeny of Blastulidium paedophthorum, a widespread brood parasite of Daphnia spp. Appl Environ Microbiol 81:5486–5496. https://doi.org/10.1128/AEM.01369-15
Edgar RC, Haas B, Clemente JC, Quince C, Knight R (2011) UCHIME improves sensitivity and speed for chimera detection. Bioinformatics 27:2194–2200. https://doi.org/10.1093/bioinformatics/btr381
Esling P, Lejzerowicz F, Pawlowski J (2015) Accurate multiplexing and filtering for high-throughput amplicon-sequencing. Nucleic Acids Res 43:2513–2524. https://doi.org/10.1093/nar/gkv107
Faure E, Not F, Benoiston AS, Labadie K, Bittner L, Ayata SD (2019) Mixotrophic protists display contrasted biogeographies in the global ocean. ISME J 13:1072–1083. https://doi.org/10.1038/s41396-018-0340-5
Ferreira de Araujo AS, Mendes LW, Lemos LN, Lopes Antunes JE, Aguiar Beserra JE Jr, do Carmo Catanho Pereira de Lyra M, do Vale Barreto Figueiredo M, de Almeida Lopes ÂC, Ferreira Gomes RL, Melgaço Bezerra W, Maciel Melo VM, de Araujo FF, Geisen S (2018) Protist species richness and soil microbiome complexity increase towards climax vegetation in the Brazilian Cerrado. Comm Biol 1:135. https://doi.org/10.1038/s42003-018-0129-0
Fiore-Donno AM, Rixen C, Rippin M, Glaser K, Samolov E, Karsten U, Becker B, Bonkowski M (2018) New barcoded primers for efficient retrieval of cercozoan sequences in high-throughput environmental diversity surveys, with emphasis on worldwide biological soil crusts. Mol Ecol Resour 18:229–239. https://doi.org/10.1111/1755-0998.12729
Fiore-Donno AM, Richter-Heitmann T, Degrune F, Dumack K, Regan KM, Marhan S, Boeddinghaus RS, Rillig M, Friedrich MW, Kandeler E, Bonkowski M (2019) Functional traits and spatio-temporal structure of a major group of soil protists (Rhizaria: Cercozoa) in a temperate grassland. Front Microbiol 10:1332. https://doi.org/10.3389/fmicb.2019.01332
Fiore-Donno AM, Richter-Heitmann T, Bonkowski M (2020) Contrasting responses of protistan plant parasites and phagotrophs to ecosystems, land management and soil properties. Front Microbiol 11:1823. https://doi.org/10.3389/fmicb.2020.01823
Fischer M, Bossdorf O, Gockel S, Hänsel F, Hemp A, Hessenmöller D, Korte G, Nieschulze J, Pfeiffer S, Prati D, Renner S, Schöning I, Schumacher U, Wells K, Buscot F, Kalko EK, Linsenmair KE, Schulze E-D, Weisser WW (2010) Implementing largescale and longterm functional biodiversity research: The Biodiversity Exploratories. Basic Appl Ecol 11:473–485. https://doi.org/10.1016/j.baae.2010.07.009
Foesel BU, Nägele V, Naether A, Wüst PK, Weinert J, Bonkowski M, Lohaus G, Polle A, Alt F, Oelmann Y, Fischer M, Friedrich MW, Overmann J (2014) Determinants of Acidobacteria activity inferred from the relative abundances of 16S rRNA transcripts in German grassland and forest soils. Environ Microbiol Rep 16:658–675. https://doi.org/10.1111/1462-2920.12162
Foley M, Deacon J (1985) Isolation of Pythium oligandrum and other necrotrophic mycoparasites from soil. T Brit Mycol Soc 85:631–639
Foster ZSL, Weiland JE, Scagel CF, Grünwald NJ (2020) The composition of the fungal and oomycete microbiome of Rhododendron roots under varying growth conditions, nurseries and cultivars. Phytobiomes J 4:156–164. https://doi.org/10.1094/PBIOMES-09-19-0052-R
Frank DN (2009) BARCRAWL and BARTAB: software tools for the design and implementation of barcoded primers for highly multiplexed DNA sequencing. BMC Bioinf 10:362. https://doi.org/10.1186/1471-2105-10-362
Garvetto A, Nézan E, Badis Y, Bilien G, Arce P, Bresnan E, Gachon CMM, Siano R (2018) Novel widespread marine Oomycetes parasitising diatoms, including the toxic genus Pseudo-nitzschia: genetic, morphological, and ecological characterisation. Front Microbiol 9:2918. https://doi.org/10.3389/fmicb.2018.02918
Geisen S, Tveit A, Clark IM, Richter A, Svenning MM, Bonkowski M, Urich T (2015) Metatranscriptomic census of active protists in soils. ISME J 9:2178–2190. https://doi.org/10.1038/ismej.2015.30
George PBL, Lallias D, Creer S, Seaton FM, Kenny JG, Eccles RM, Griffiths RI, Lebron I, Emmett BA, Robinson DA, Jones DL (2019) Divergent national-scale trends of microbial and animal biodiversity revealed across diverse temperate soil ecosystems. Nat Commun 10:1107. https://doi.org/10.1038/s41467-019-09031-1
Gómez-Aparicio L, Ibáñez B, Serrano MS, De Vita P, Àvila JM, Pérez-Ramos IM, García LV, Sánchez ME, Marañón T (2012) Spatial patterns of soil pathogens in declining Mediterranean forests: implications for tree species regeneration. New Phytol 194:1014–1024. https://doi.org/10.1111/j.1469-8137.2012.04108.x
Hall TA (1999) BioEdit: a user-friendly biological sequence alignment editor and analysis program for Windows 95/98/NT. Nucleic Acids Symp Ser 41:95–98
Hayden KJ, Hardy GESJ, Garbelotto M (2013) Oomycetes diseases. In: Gonthier P, Nicolotti G (eds) Infectious forest diseases. CAB International, Wallingford, pp 518–545
Jassey VE, Lamentowicz M, Bragazza L, Hofsommer ML, Mills RTE, Buttler A, Signarbieux C, Robroek BJM (2016) Loss of testate amoeba functional diversity with increasing frost intensity across a continental gradient reduces microbial activity in peatlands. Eur J Protistol 55:190–202. https://doi.org/10.1016/j.ejop.2016.04.007
Kaiser K, Wemheuer B, Korolkow V, Wemheuer F, Nacke H, Schöning I, Schrumpf M, Daniel R (2016) Driving forces of soil bacterial community structure, diversity, and function in temperate grasslands and forests. Sci Rep 6:33696. https://doi.org/10.1038/srep33696
Katoh K, Standley DM (2013) MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol 30:772–780. https://doi.org/10.1093/molbev/mst010
Kramer S, Dibbern D, Moll J, Hünninghaus M, Koller R, Krüger D, Marhan S, Urich T, Wubet T, Bonkowski M, Buscot F, Lueders T, Kandeler E (2016) Resource partitioning between bacteria, fungi, and protists in the detritusphere of an agricultural soil. Front Microbiol 7:1524. https://doi.org/10.3389/fmicb.2016.01524
Lara E, Belbahri L (2011) SSU rRNA reveals major trends in oomycete evolution. Fungal Divers 49:93–100. https://doi.org/10.1007/s13225-011-0098-9
Lebeda A, Spencer-Phillips PTN, Cooke BM (2008) The downy mildews - genetics, molecular biology and control. Springer, Netherlands. https://doi.org/10.1007/978-1-4020-8973-2
Lentendu G, Mahé F, Bass D, Rueckert S, Stoeck T, Dunthorn M (2018) Consistent patterns of high alpha and low beta diversity in tropical parasitic and free-living protists. Mol Ecol 27:2846–2857. https://doi.org/10.1111/mec.14731
Liebe S, Wibberg D, Winkler A, Pühler A, Schlüter A, Varrelmann M (2016) Taxonomic analysis of the microbial community in stored sugar beets using highthroughput sequencing of different marker genes. FEMS Microbiol Ecol 92:92. https://doi.org/10.1093/femsec/fiw004
Lievens B, Hanssen IRM, Vanachter ACRC, Cammue BPA, Thomma BPHJ (2004) Root and foot rot on tomato caused by Phytophthora infestans detected in Belgium. Plant Dis 88:86. https://doi.org/10.1094/PDIS.2004.88.1.86A
Lifshitz R, Hancock JG (1983) Saprophytic development of Pythium ultimum in soil as a function of water matric potential and temperature. Phytopathology 73:257–261
Löbmann MT, Vetukuri RR, de Zinger L, Alsanius BW, Grenville-Briggsa LJ, Walter AJ (2016) The occurrence of pathogen suppressive soils in Sweden in relation to soil biota, soil properties, and farming practices. Appl Soil Ecol 107:57–65. https://doi.org/10.1016/j.apsoil.2016.05.011
Lorang J (2019) Neotrophic exploitation and subversion of plant defense: a lifestyle or just a phase, and implications in breeding resistance. Phytopathology 109:332–345. https://doi.org/10.1094/PHYTO-09-18-0334-IA
Marano AV, Jesus AL, De Souza JI, Jerônimo GH, Gonçalves DR, Boro MC, Rocha SCO, Pires-Zottarelli CLA (2016) Ecological roles of saprotrophic Peronosporales (Oomycetes, Straminipila) in natural environments. Fungal Ecol 19:77–88. https://doi.org/10.1016/j.funeco.2015.06.003
Michu E, Mráčková M, Vyskot B, Žlůvová J (2010) Reduction of heteroduplex formation in PCR amplification. Biol Plant 54:173–176
Nacke H, Thürmer A, Wollherr A, Will C, Hodac L, Herold N, Schöning I, Schrumpf M, Daniel R (2001) Pyrosequencing-based assessment of bacterial community structure along different management types in German forest and grassland soils. PLoS ONE 6:e17000. https://doi.org/10.1371/journal.pone.0017000
Oksanen J, Blanchet FG, Kindt R, Legendre P, Minchin PR, O’Hara RB, Simpson GL, Solymos P, Stevens MHH, Wagner H (2013) Vegan: community ecology package. R Package version 2:10. http://CRAN.R-project.org/package=vegan. Accessed March 2020
Packer A, Clay K (2000) Soil pathogens and spatial patterns of seedling mortality in a temperate tree. Nature 404:278–281. https://doi.org/10.1038/35005072
Pandaranayaka EPJ, Frenkel O, Elad Y, Prusky D, Harel A (2019) Network analysis exposes core functions in major lifestyles of fungal and oomycete plant pathogens. BMC Genomics 20:1020. https://doi.org/10.1186/s12864-019-6409-3
Précigout P-A, Claessen D, Makowski D, Robert C (2020) Does the latent period of leaf fungal pathogens reflect their trophic type? A meta-analysis of biotrophs, hemibiotrophs, and necrotrophs. Phytopathology in press 110:345–361. https://doi.org/10.1094/PHYTO-04-19-0144-R
R Development Core Team (2014) R: a language and environment for statistical computing. Austria, Vienna
Riit T, Tedersoo L, Drenkhan R, Runno-Paurson E, Kokko H, Anslan S (2016) Oomycete-specific ITS primers for identification and metabarcoding. MycoKeys 14:17–30. https://doi.org/10.3897/mycokeys.14.9244
Rognes T, Flouri T, Nichols B, Quince C, Mahé F (2016) VSEARCH: a versatile open source tool for metagenomics. PeerJ 4:e2584. https://doi.org/10.7717/peerj.2584
Sapkota R, Nicolaisen M (2015) An improved high throughput sequencing method for studying oomycete communities. J Microbiol Methods 110:33–39. https://doi.org/10.1016/j.mimet.2015.01.013
Sapp M, Ploch S, Fiore-Donno AM, Bonkowski M, Rose LE (2018) Protists are an integral part of the Arabidopsis thaliana microbiome. Environ Microbiol Rep 20:30–43. https://doi.org/10.1111/1462-2920.13941
Sapp M, Tyborski N, Linstädter A, López Sánchez A, Mansfeldt T, Waldhoff G, Bareth G, Bonkowski M, Rose LE (2019) Site-specific distribution of oak rhizosphere-associated oomycetes revealed by cytochrome c oxidase subunit II metabarcoding. Ecol Evol 9:10567–10581. https://doi.org/10.1002/ece3.5577
Savory F, Leonard G, Richards TA (2015) The role of horizontal gene transfer in the evolution of Oomycetes. PLoS Path 11:e1004805. https://doi.org/10.1371/journal.ppat.1004805
Schlatter D, Kinkel L, Thomashow L, Weller D, Paulitz T (2017) Disease suppressive soils: new insights from the soil microbiome. Phytopathology 107:1284–1297. https://doi.org/10.1094/PHYTO-03-17-0111-RVW
Schloss PD, Westcott SL, Ryabin T, Hall JR, Hartmann M, Hollister EB, Lesniewski RA, Oakley BB, Parks DH, Robinson CJ, Sahl JW, Stres B, Thallinger GG, Van Horn DJ, Weber CF (2009) Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl Environ Microbiol 75:7537–7541. https://doi.org/10.1128/AEM.01541-09
Singer D, Lara E, Steciow MM, Seppey CVW, Paredes N, Pillonel A, Oszako T, Belbahri L (2016) High-throughput sequencing reveals diverse oomycete communities in oligotrophic peat bog micro-habitat. Fungal Ecol 23:42–47. https://doi.org/10.1016/j.funeco.2016.05.009
Spanu PD, Panstruga R (2017) Editorial: Biotrophic plant-microbe interactions. Front Plant Sci 8:192. https://doi.org/10.3389/fpls.2017.00192
Steciow MM, Lara E, Paul C, Pillonel A, Belbahri L (2014) Multiple barcode assessment within the Saprolegnia-Achlya clade (Saprolegniales, Oomycota, Straminipila) brings order in a neglected group of pathogens. IMA Fungus 5:439–448. https://doi.org/10.5598/imafungus.2014.05.02.08
Thines M (2014) Phylogeny and evolution of plant pathogenic oomycetes—a global overview. Eur J Plant Pathol 138:431–447. https://doi.org/10.1007/s10658-013-0366-5
Thines M, Choi Y-J (2016) Evolution, diversity, and taxonomy of the Peronosporaceae, with focus on the genus Peronospora. Phytopathology 106:6–18. https://doi.org/10.1094/PHYTO-05-15-0127-RVW
Thines M, Kamoun S (2010) Oomycete–plant coevolution: recent advances and future prospects. Curr Opin Plant Biol 13:427–433. https://doi.org/10.1016/j.pbi.2010.04.001
Venter PC, Nitsche F, Domonell A, Heger P, Arndt H (2017) The protistan microbiome of grassland soil: diversity in the mesoscale. Protist 168:546–564. https://doi.org/10.1016/j.protis.2017.03.005
Zhao H, Li X, Zhang Z, Zhao Y, Chen P, Zhu Y (2018) Drivers and assemblies of soil eukaryotic microbes among different soil habitat types in a semi-arid mountain in China. PeerJ 6:e6042. https://doi.org/10.7717/peerj.6042
Acknowledgments
We are very grateful to Linhui Jiang and Christopher Kahlich for invaluable help in the lab. We thank Graham Jones, Scotland, for writing the R scripts selecting barcodes. At the University of Geneva (CH), we thank Jan Pawlowski and Emanuela Reo and we are liable to the Swiss National Science Foundation Grant 316030 150817 funding the MiSeq instrument. We thank the managers of the Exploratories, Swen Renner, Kirsten Reichel-Jung, Kerstin Wiesner, Katrin Lorenzen, Martin Gorke, Miriam Teuscher and all former managers for their work in maintaining the plot and project infrastructure; Simone Pfeiffer and Christiane Fischer for giving support through the central office; Jens Nieschulze and Michael Owonibi for managing the central database; and Markus Fischer, Eduard Linsenmair, Dominik Hessenmüller, Daniel Prati, Ingo Schöning, François Buscot, Ernst-Detlef Schulze, Wolfgang W. Weisser and the late Elisabeth Kalko for their role in setting up the Biodiversity Exploratories project. Fieldwork permits were issued by the responsible state environmental offices of Baden-Württemberg, Thüringen and Brandenburg.
Funding
Open Access funding enabled and organized by Projekt DEAL. This work was funded by the DFG Priority Program 1374 “Infrastructure-Biodiversity-Exploratories”, subproject BO 1907/18-1 (PATHOGEN) to MB and AMFD.
Author information
Authors and Affiliations
Contributions
Conceptualisation of the PATHOGEN project and interpretation of the data (Bonkowski and Fiore-Donno). Amplifications, Illumina sequencing, bioinformatics pipeline, statistical analyses and first draft of the manuscript (Fiore-Donno). Funding acquisition, revisions of the manuscript (Bonkowski).
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethics approval
Not applicable.
Consent to participate
Not applicable.
Consent for publication
Not applicable.
Code availability
Not applicable.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fiore-Donno, A.M., Bonkowski, M. Different community compositions between obligate and facultative oomycete plant parasites in a landscape-scale metabarcoding survey. Biol Fertil Soils 57, 245–256 (2021). https://doi.org/10.1007/s00374-020-01519-z
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00374-020-01519-z