Abstract
Segmentation of anatomical structures on 2D images of cardiac exams is necessary for performing 3D volumetric analysis, enabling the computation of parameters for diagnosing cardiovascular disease. In this work, we present robust algorithms to automatically segment cardiac imaging data and generate a volumetric anatomical reconstruction of a patient-specific heart model by propagating active contour output within a patient stack through a self-supervised learning model. Contour initializations are automatically generated, then output segmentations on sparse image slices are transferred and merged across a stack of images within the same heart data set during the segmentation process. We demonstrate whole-heart segmentation and compare the results with ground truth manual annotations. Additionally, we provide a framework to represent segmented heart data in the form of implicit surfaces, allowing interpolation operations to generate intermediary models of heart sections and volumes throughout the cardiac cycle and to estimate ejection fraction.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Cardiovascular disease continues to rank among the leading causes of global mortality, highlighting the increasing urgency surrounding the need for diagnosis, treatment, and preventive measures concerning cardiac health [50]. To evaluate patient well-being, healthcare providers use imaging methods such as computed tomography (CT) and magnetic resonance imaging (MRI), which are considered the gold standard for cardiac imaging [15]. These imaging methods generate information in the form of three-dimensional volumetric data sets of the anatomical structures and vasculature of the heart, including cardiac chamber morphology and coronary artery angiography, using high-resolution cross-sectional image acquisition techniques. The data sets are then interpreted to find abnormalities, such as morphological aberrations, aneurysms, and cardiac chamber enlargement. Functional information such as ejection fraction or blood flow measurements related to the myocardium and cardiac chambers can be performed using volumetric data analysis by comparing the imaging data acquired at different phases of the cardiac cycle. Subsequent analysis of these exams allows an early and accurate evaluation of cardiac function which is vital in treating heart disease.
Performing volumetric analyses of anatomical structures requires segmenting these structures of interest on sectional 2D digital images. This segmentation process is achieved by delineating areas of interest, such as the cardiac chambers, to separate them from the rest of the organ structures. This allows for better characterization and analysis of structures embedded within background body parts. These segmentations can encompass MPRs (multiplanar reformations) in various body planes such as the coronal, axial, and sagittal planes or dedicated cardiac imaging planes such as the horizontal long axis (4 chamber view), vertical long axis (2 chamber view), and short axis view for use in visual assessment by radiologists. A full-volume segmentation of these structures allows users to generate 3D reconstructions separated from background tissues. Multi-parametric analysis can be performed to detect normal and abnormal states of these organs and tissues, alongside generating 4D information and functional analysis over the cardiac cycle.
In the past, medical professionals manually segmented patient exams to capture regions of interest, which was time- and resource-intensive. As computing technology improved, many assistive software tools [23] have been developed for automatic data post-processing. However, despite annual advances, segmentation of anatomical structures in diagnostic images remains difficult; cardiac segmentation remains an active problem in medical image processing [17] due to limitations of scanner technology and technical challenges of imaging human subjects, particularly with cardiac imaging, resulting in suboptimal data acquisition and image artifacts. Accurate visualization, simulation, and animation of anatomical structures are crucial for medical professionals in diagnosing cardiovascular diseases [32]. Cardiac imaging data are unique in contrast to other imaged organs, such that the inclusion of a temporal domain adds significant value in analyzing physiological parameters such as the ejection fraction and visualization of muscle contraction across the spatial domain; the need for analysis throughout the entire cardiac cycle further highlights the importance of animation and how additional information provided and presented in 4D data can be instrumental in patient care. Among different representations of these 4D spatiotemporal data, implicit surface representation is well suited for acquired heart data in animation and volumetric computations. These implicit functions offer a versatile and compact representation that encodes the geometry and the temporal change in the geometry based on level sets. This allows for efficient storage, manipulation, and representation of complex 3D structures without explicitly storing surface geometry.
An implicit representation allows users to interpolate between keyframes to fill in or interpolate missing information. Implicit functions can also be used to easily test whether a point is inside or outside a volume, making it a prime candidate for representation for easy computation of volume, ejection fraction, and discretization.
Our work presents a robust algorithm that can automatically segment cardiac structures from patient data, developed using an actual patient exam. We minimize user input by automatically generating initial segmentations through image processing techniques and morphological geodesic active contour models [34]. We present a process to automatically generate these inputs, though they can also take in inputs from other segmentation frameworks. These initial segmentations serve as initializers for a self-supervised annotation propagation model [51] trained on cardiac data to propagate initial segmentations for target structures of interest throughout the volume. New structures of interest are detected as the pipeline progresses throughout the sections and are added to the propagation module to assist in segmentation. We compared our output segmentations to ground truth segmentations performed by a medical professional. Furthermore, we present a technique to represent segmented volumes as implicit surfaces, where input image or volume data can be interpolated spatially or temporally to compute geometric and temporal data for visualization and animation. This representation can also estimate ejection fraction metrics from reconstructed left ventricular models.
1.1 Main contributions
Our main contributions are as follows.
-
1.
We propose a robust, end-to-end, automatic edge-based contouring for segmentation that generates structure outlines to extract major cardiac structures (chambers and great vessels) from exam data sets. This contouring can provide segmentations standalone and optionally supplement existing segmentation frameworks by iterating from their outputs.
-
2.
We reconstruct a patient-specific 3D and/or 4D heart model from the computed and propagated contours for modeling and visualization.
-
3.
We provide a framework extending implicit surface work to represent 3D and 4D cardiac volumes. We introduce additional boundary conditions to help regularize and facilitate clean and accurate interpolation without self-intersection. This allows for representing each chamber with an independent implicit function and generating new intermediary outputs by interpolating and reconstructing between spatial and temporal keyframes for accurate dynamic motion of imaged cardiac structures.
-
4.
From our presented implicit surface framework, we can also estimate cardiac volumetric measurements, such as the ejection fraction.
2 Related work
Much work has been done on segmenting cardiovascular anatomy throughout the years and has been extensively covered in reviews [17, 42]. These techniques can be classified into three approaches: edge-based, model-based, and learning-based [9, 13].
Edge-based techniques in segmentation use active contour models, or snakes, to guide a level set toward a region of interest [19]. Active contour models were first introduced in 1988 [22] and have become a staple in classical image segmentation [43]. Many variations of the active contour model have developed over time. The Mumford–Shah model [33] uses a variational approach by minimizing energy functionals for piecewise constant functions. Geodesic active contours [7] combine image information and geometric properties of evolving curves to capture boundaries using geodesic distance. Chan-Vese contours [8] utilize both the regional mean intensity and its complement in the image to segment objects with relatively homogeneous intensities. Morphological geodesic active contours [34] expand on the work of Caselles et al. [7] by substituting PDEs with morphological operators for robustness to noise.
Model-based techniques often revolve around atlas-based segmentation methods, which require matching image features with deformable registration methods [6, 21]. These techniques rely on prior information and require multiple forms of pre-built annotated heart databases, which can be registered using tools like elastix [24]. These models can output high-quality segmentations but may be susceptible to large shape variability. Additionally, the construction of manually annotated heart atlases may not be practical due to the time-intensive work needed to create annotations. Atlas-based registration based on unsupervised deep learning techniques [3] also requires a significant amount of labeled data to fit a network and may not adjust well to exams that differ from the training distribution.
Deep learning segmentation techniques have been a dominant presence in medical applications. The explosion of convolutional neural networks for use in image-based learning tasks [25] and U-Nets [20, 40] has bolstered image-based learning methods in medical applications, including cardiac segmentation [10, 18]. These learning-based frameworks have found rapid growth and success in segmentation work, particularly in broader medical applications. Recent works have achieved good results in related applications, such as breast cancer segmentation of postmastectomy patients [28, 29] and multi-organ segmentation in male pelvis CT scans [41] which can be used to supplement and enhance the workflow of radiologists.
However, these networks are heavily based on data; these methods require large amounts of labeled data to train. Though public challenge data sets [45] are becoming increasingly available for use [14], comprehensive annotated human cardiac data is very sparse. For optimal accuracy, these online data sets use manual segmentations from medical specialists, which require significant time commitments and collaboration to generate. Obtaining many quality human heart annotations remains difficult and labor-intensive; moreover, many of these data sets focus on identifying features of patients with irregular conditions, such as exams with cancerous growths, lesions, and calcium buildup. These features would generally be atypical of a healthy patient and can make it hard to generalize outside the scope of the data set. Trained models may also face difficulties in generalizing to data outside their training parameters, such as exams taken from different scanners or vendors [9]. The vast number of structures of interest within a patient exam also makes the problem of whole-heart segmentation using classical deep learning segmentation techniques difficult, as many public challenge data sets only focus on providing labels for one or two structures. Additionally, the intermediary process in training may resemble black boxes, and it can be challenging to quantify the origins of a network’s performance or behavior.
This scarcity of labels and data privacy/ethics laws can also heavily limit the practical application of many deep learning methods. As a result, approaches with classical image processing [36], active contouring [1, 37], and a combination of both [2, 27] are often used instead for simple segmentation tasks. However, recent trends have found some success in auto-segmentation [16], unsupervised learning methods, and semi-supervised methods for medical computing. Recent developments in self-supervised learning techniques [51] allow for the propagation of segmentations across an input volume but are not fully automatic. Although the propagation framework is robust, it requires an annotation for each region of interest to create labels along volume slices, and the propagation framework can only propagate the structures provided in the initial manual annotation. This issue is further compounded within the context of cardiac data, where simply providing a single annotated section is insufficient, as a single patient exam has many structures of interest interspersed between the sections, which may not always be present at a given initial index. This method would then require a new set of annotations each time a new structure of interest appears in a section. While manual annotations for initialization are difficult to find, since our work automates this computation, we can use the above annotation propagation system. In other words, we generate automatic initial segmentations of blood vessels and chambers of the heart for active contouring operations and use a self-supervised learning-based propagation model [51] in a limited manner to complete missing data and to ensure consistency across labels. Our hybrid method for automatic segmentation, annotation computation, and propagation is explained in Sects. 3.3.1 and 3.4.
In medical visualization, proper visual representation and rendering of patient heart anatomy are essential for VR, AR, educational, and surgical applications [5]. Classic representations of medical images involve volume data rendered using raycasting [52], tetrahedralization [26], or after isosurface extraction [30, 36]. In shape transformation, deep learning methods have been proposed [39, 47] to represent geometries in a latent space to generate and morph models by interpolating the latent space parameters. However, these works focus on representing and morphing between styles, rather than animating and measuring geometric quantities. These networks also require significant amounts of complex training data.
In dynamic cardiac data representation, very little work, if any, has been done; shape transformation in this topic remains a promising avenue. Turk and O’Brien [49] introduced implicit functions to smoothly transform 2D or 3D manifold shapes to another. Since individual cardiac chambers and the outside heart structure can be represented as genus-0 manifolds (excepting the vascular connections) and are functionally dynamic (moving), we propose a 4D (spatiotemporal) implicit function for dynamic heart representation and analysis. We also use this representation to estimate cardiac parameters such as the ejection fraction.
3 Method
3.1 Overview
We present a brief overview of our methodology. Once data are collected (Sect. 3.2) and preprocessed (Sect. 3.2.1), we automatically generate the initial level sets for each contour using distance map operations (Sect. 3.3.2) and the watershed algorithm (Sect. 3.3.3). These contour models then iteratively grow to capture regions of interest near their initializations in each image section. The output of these contour models is then used as input for the propagation model in Sect. 3.4, which propagates the contours across the volume sections. This process relates contours between slices that correspond to the same substructure of the heart and can also serve as initializations, similar to the distance map and watershed operations. Further processing occurs to account for noisy contours and outliers.
Once segmented substructures are provided, we fit implicit functions (Sect. 3.5), one for each substructure. We ensure that these implicit surfaces do not intersect each other using appropriate boundary conditions (Sect. 3.5.2). Lastly, we detail ejection fraction estimation in Sect. 3.5.4 with implicit representation.
We additionally define the terminology used throughout our work to refer to our data. We denote segmentation as dividing or creating portions of an image or volume data set into masks defining regions of interest. The contours seen throughout our process serve as segmentations of structures of interest from the surrounding tissue and other structures, where the boundaries of the curves represent the border of a structure and the interiors of the curve represent areas belonging to that structure of interest. Annotations refer to data often manually drawn as ground truth images in public data sets. These annotations consist of contours for segmentation and additional metadata, such as the names or identifiers of the segmented regions, also called labels, data collection device information, etc.
3.2 Data
We utilize both CT and MRI data sets to evaluate our work. For our CT data set, we used a set of de-identified cardiac CT volume images stored in DICOM format with 512 \(\times \) 512 pixels in the axial direction with a slice thickness of 0.8 mm. This volume contains 354 image sections, with a voxel size of 0.38 \(\times \) 0.38 \(\times \) 0.40 mm. These exams were taken on a Philips iCT 256-slice scanner with 35% contrast dye. To evaluate the performance of our results for this data set, we obtained the ground truth labels in each section for the core anatomical substructures. This ground truth is of the same dimensions as its corresponding data and was manually annotated by a cardiovascular specialist (see Acknowledgments). Annotations were created for the arteries, including the aorta, superior vena cava, pulmonary veins, atria, and ventricles. Each structure was labeled with an integer identifying the segmentation as belonging to a specific structure.
For evaluation of MRI data, 4D MRI images from the 2017 MICCAI Automated Cardiac Diagnosis Challenge (ACDC) public data set [4], MM-WHS MRI data set [53], and Sunnybrook data set [38] were used. Each volume in the data set contains an equivalent corresponding ground truth volume containing relevant ventricular segmentations.
3.2.1 Preprocessing
We define our image data set as an ordered list \(\mathcal {D} = (I_{1}, \dots , I_{n})\), consisting of n image slice elements. Each image \(I_{i}: [0, X] \times [0, Y] \rightarrow \mathbb {R}^{+}\) is associated with a set of binary images \(\mathcal {U}_{i}\), which is initially empty, but will contain binary images \(u_{i1},..., u_{im}\) representing segmentations within the image \(I_{i}\). The edges of these binary segmentations serve as contours to delineate the heart substructures.
To standardize values, we convert each image \(I_{i}\) of the CT data to the Hounsfield scale to compute the radiodensity of each pixel. As structures of interest will be highlighted by contrast dye, in order to simplify the image feature search space, we threshold the image section using Hounsfield units and create a mask to remove pixels corresponding to fat, air, and high-density bone values. This thresholded output mask may contain a few specks in the fat/tissue regions. Additionally, a few pixels or specks may be thresholded out of some structures within the mask. As structures of interest within cardiac data (such as vessels) tend to be approximate smooth, contiguous genus-0 topology, we want to ensure these components do not contain holes or internal specks. To ensure consistency, we apply morphological operations such as closing and opening to remove and fill these small artifacts. From our observations, these operations do not incur any meaningful loss of information. In the following sections, we process this mask further to generate initializations for contouring operations.
3.3 Morphological geodesic active contours
Classical contour-based algorithms are a staple of image-based biomedical segmentation tasks, but have drawbacks. Active contours are highly sensitive to initial configurations and noise. When applying active contours on image data sets, noise may be mistaken for small gradients in the image. This can directly affect the results and create less-than-ideal segmentation output.
We use active geodesic morphological contours [34] to overcome this problem. This implementation uses morphological operators over traditional partial differential equations, offering more robustness to noise and better handling of ambiguous boundaries, which are common features of medical data. This allows for faster and more stable convergence. Typically in active contour applications, the user must provide an input segmentation to initialize the contour. We avoid this problem by automatically generating well-defined initializations (Sect. 3.3.1). We also ameliorate the problem of contours getting stuck in local minima by using a positive balloon force [11] to grow from inside the cardiac substructures. This positive balloon force also accounts for the general convexity of most cardiac substructures, allowing contours to dilate and inflate contour curves to envelope regions of interest without over-segmentation.
Implementing morphological geodesic active contours can be viewed as the sum of three components: smoothing force, balloon force, and image attraction force [34]. These three components can be rewritten as morphological equations that use binary erosion and binary dilation to act upon some image to evolve the contour toward nearby edges. Smoothing force is first applied to smoothen jagged lines. Balloon force helps guide the contour out of uninformative homogeneous regions and avoids cases of the contour getting stuck in local minima. Finally, attraction force serves as an extrinsic operator that acts on the contour to evolve the curve, pulling it toward edge features within the image. This attraction force relies on an image edge indicator function g(I), which is lower at feature boundaries and higher in homogeneous regions of the image. In the following section, we describe our methods to setup these parameters for the active contour algorithm.
3.3.1 Contour initialization
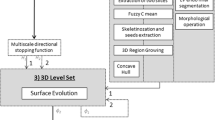
Each morphological geodesic active contours operation requires an initial contour and an input image g(I), preprocessed to highlight potential contours and edges. The initial segmentation serves as the starting point of the contour and grows to envelop a region in g(I). Typical applications would prompt the user to manually draw an enclosed curve around a region of interest to initialize a starting segmentation [31]. However, we do not require user input for this process; instead, we automatically generate initial starting shapes for the contours in each section, which is a rough segmentation of a region of interest that grows or shrinks as the contours iterate using the active contour algorithm. A pipeline of our segmentation process is shown in Fig. 1.
We use the preprocessed thresholded image (output of Sect. 3.2.1) as our g(I). This image is processed to create a Gaussian gradient magnitude image, where values in the gradient image increase when approaching a change in the image or an edge and approaches zero when on the edge itself. This is done by computing histogram equalization on the image, using the gradient magnitude operator to highlight edges, and applying a Gaussian filter block \(G_\sigma \) with \(\sigma = 5\) to reduce noise within substructures while still preserving edge information. Figure 2 shows a sample morphological geodesic active contour operation with the Gaussian gradient magnitude image.
For the initialization of contours, any set of binary initializations can be provided, whether manually drawn or automatically generated from our framework or other segmentation algorithms. For our automatic implementation, we combine two methods, Euclidean distance map transform (Sect. 3.3.2) and the watershed algorithm (Sect. 3.3.3), to create these inputs to the geodesic active contour algorithm. These contours are then propagated across the volume using an unsupervised network (Sect. 3.4).
3.3.2 Euclidean distance map
Evolution of a geodesic active contour curve on an input image (left). The curve evolution is shown over the Gaussian gradient magnitude image (right). The Gaussian gradient magnitude operation highlights strong edges in the input image and is used to help the contour grow and capture the shape of the aorta
First three images depict a distance transform (2) on the input image (1) and its corresponding connected components (3). The fourth and fifth pictures depict markers for the watershed seed generation (4) computed from the centroids in (4) and the corresponding watershed output segmentations (5). The watershed output (5) captures the full regions of interest for the green and yellow regions, creating a better initialization than in (3). However, the red marker has over-segmented and enveloped the rest of the section due to factors such as noise. This marker will be handled by growing a contour from the distance map component. The last image (6) depicts a merging of valid outputs from watershed (5) and rest of the output from the distance map operation (3)
Our first set of initial segmentations is done by retrieving a thresholded distance map (output of Sect. 3.2.1) of the image section and finding its connected components. We binarize the thresholded image from Sect. 3.2.1 and apply a Euclidean distance transform to generate our initial segmentations. This generates a corresponding distance-transformed image containing the squared distance values for each pixel to the nearest 0 pixels. We then threshold at the 20th percentile value and binarize this distance-transformed image. This operation has the benefit of retaining regions closest to the center of structures with a lot of surrounding information. It also reduces noise and separates substructures that may be too close to each other and would otherwise be seen as a single connected component. From this transformation, we generate a set of connected components \(\{u_{1}, \dots , u_{k}\}\) from contiguous 1-pixels as initial segmentations and append them to the set of initial segmentations for the current section \(\mathcal {U}_{i}\). A sample distance map transforms, and its connected components are depicted in Fig. 3. After computing the initial segmentations using the distance map, we compute additional initializations using the watershed algorithm.
3.3.3 Watershed
The initial segmentations computed from the distance map transform in Sect. 3.3.2 may not encompass the entire region we want to fully segment. Rather, they may only capture an adequately sized portion of the desired area to iterate on. Active contours often completely envelop the target region when iterating upon these initializations. However, for areas with high variance in density values (e.g., calcification in the blood vessels) or with noisy, ambiguous edges and interior values (e.g., lower heart chambers and sections of the aorta), the initial segmentation done by the distance map transform may be insufficient and small, leading to the active contour algorithm getting stuck in local minima, especially if there is noise or artifacts in the surrounding region of these small contours. Annealing operations can be performed but at great computational cost; instead, we introduce the watershed algorithm [35] as a simpler way to address this problem.
Compared to the distance map transform, watershed segmentations tend to capture significantly more of the structures of interest and are more suited for regions that are non-convex and high in variance. Since most of the structure of interest will be captured in the watershed initializations, active contour operations on these require significantly fewer iterations and less computation time. These operations will be more likely to grow to encompass the entire structure of interest, if not already done. However, the initializations generated from watershed may be prone to over-segmentations, which can cause the output to flow over the region of interest and overtake a large portion of the image slice. Such cases are trivial to identify and correct and can be easily removed. A result of the watershed operation is shown in Fig. 3.
To compute the watershed initializations, we start from the connected components computed in Sect. 3.3.2. We calculate the centroids of each connected component and set an initial basin of 1 pixel at each centroid coordinate for a watershed operation. We then retrieve the delineated regions and remove outliers. Incorrect segmentations are trivial to detect: we remove low-quality segmentations that are excessively large or small, and append the remaining segmentations to the set \(\mathcal {U}_{i}\).
3.3.4 Final image segmentation using active contours
After computing the initializations, we use active contours to iteratively grow from the initial segmentations until a stopping criterion is reached. We apply this active contour method on the Gaussian gradient magnitude image g(I). This is done for every image section in the volume.
During iteration, there may be instances in which contours overlap with each other contours. This may be due to the initializations being placed in the same general area, roughly textured regions of interest creating uneven distributions of initializations (e.g., occasionally seen in the right atrium), or long and narrow areas of interest (e.g., pulmonary arteries). To resolve these overlaps, contours that sufficiently overlap each other according to their Sørensen–Dice coefficient [12, 44] are merged. Outliers that are too large or too small are discarded, and the resultant segmentations for the current section are used as input to propagate throughout the volume as described in the following section.
3.4 Contour propagation with correspondence networks
After computing all the contours of an image as explained in Sect. 3.3.4, we apply an unsupervised learning model to propagate these contours throughout the volume sections.
For the propagation module, we utilize the Sli2Vol correspondence network detailed in [51]. One limitation raised with the original Sli2Vol work was its need for an input annotated slice to propagate information properly. The tendency of cardiac data, particularly CT volumes, to present structures of interest at differing section indices within the volume poses a problem, as all of the structures we want to segment may not be present in a single initial section. For example, at the beginning of a CT scan, only the blood vessels such as the aorta are present, which will then taper off into other structures or arteries further down the sections; other interesting structures of interest such as the chambers may not be present until halfway or at the end of the volume sections. The Sli2Vol framework can propagate only segmentations for structures of interest that have already been provided in an input section. To account for additional structures throughout the sections, a new set of segmentations must be provided each time a new structure of interest presents itself.
Using our previously iterated contour outputs as input allows us to take advantage of the automatically captured structures of interest within a given section. This allows us to repeat this process for other sections to add new structures as we progress down the volume. The contours propagated down the sections can supplement the segmentation process and serve as additional initial segmentations for active contours. This also has the benefit of keeping the labels consistent throughout the volume, carrying over labels that identify specific substructures between sections.
While the Sli2Vol correspondence network training was designed for general propagation across various medical segmentation tasks, we focus solely on modalities for cardiac imaging for our work. We adopt similar training steps in [51] to output and train the model in the original work. The network is trained for each model with image augmentation applied to the training data. Each model is trained using an Adam optimizer at a 1e−4 learning rate with 4 epochs.
3.5 Implicit surface representation
Once all the images in the stack are processed and all contours computed, we represent the contours across the volume in implicit surface form, as introduced in [49]. This has multiple advantages. In storage and memory, shapes can be stored as a collection of weights within a linear system. Additionally, representing geometry in implicit form also allows us to generate interpolated outputs between sets of input keyframes to help fill in missing information, both in the spatial domain between slices and the temporal domain between 3D shapes for dynamic modeling. From this, we can retrieve intermediary states of a heart between time steps in the cardiac cycle and use them for animation or analysis. These interpolated outputs are discrete volumes that can be used in existing rendering pipelines. This framework is not limited to interpolating between 3D models in time steps; it can also interpolate between 2D sections to generate intermediary 2D sections within a 3D model. We introduce a shape transformation framework to represent our outputs as implicit surfaces.
3.5.1 Definition
Turk and O’Brien [49] defines implicit surface representation in the context of a linear system, composed of a set of k constraints, \(c_1,..., c_k \in C\), a thin-plate spline function \(\phi \), and an interpolation function f(x).
Each constraint \(c_i \in C\) is made up of up to four values (depending on the dimensionality of the data), \(c_i^x, c_i^y, c_i^z, c_i^t\), representing the location of the constraint in the x-axis, y-axis, z-axis, and timestep, respectively.
The thin-plate spline function \(\phi _{ij}\) between two constraints \(c_i, c_j \in C\) is additionally used to measure some form of similarity between two constraints and is defined as \(\phi \) in Eq. (1).
An interpolation function f(x) to solve for intermediary output is defined in Eq. (2). In this function, we solve for \(d_j\), the weights within the function, and the coefficients of P(x), a degree-one polynomial that accounts for the linear and constant portions of f. To solve for \(d_j\), the right side of the equation for \(f({\textbf {c}}_i)\) is substituted for the constraints \(h_i = f({\textbf {c}}_i)\) to get the following equation. While [49] defines \(h_i\) as a binary value of heightmap 0 or 1, we define \(h_i\) as the distance of the constraint from the nearest boundary, where a positive value denotes the constraint as within a boundary, and a negative value indicates that the constraint is outside of a boundary. A zero value indicates the constraint to be on the boundary.
Equation (2) can be represented in matrix form. Equation (3) depicts this as a linear equation for 4D data.
3.5.2 Constraints
Implicit surface representation first requires computing constraint points. In [49], the authors present two types of constraints: boundary and exterior normal constraints. Boundary constraints are computed by collecting points on the boundary of an input volume; normal constraints are found by displacing the boundary points one pixel across their normal vector. The authors approach boundary constraint computation by finding every boundary pixel. However, doing this will collect many unnecessary points and may significantly slow computation time. Therefore, for our implementation, we utilize the Teh-Chin chain approximation algorithm [48] to find the most dominant boundary points. We computed the exterior normal constraints by displacing the boundary points one unit across the vector calculated using the Sobel filter at the boundary point’s location.
However, utilizing only boundary and normal constraints from the current section may result in inaccurate interpolations that contain artifacts within the output. This is because the output attempts to fulfill local minima within the system that, although technically correct, are undesirable. Some examples are shown in Fig. 4. We solve this issue by introducing three potential types of constraints as additional regularizers, as shown in Fig. 5.
Outputs of interpolation are compared between using and not using additional constraints. Each row depicts a different left ventricular section. On the left column, outputs with artifacts are depicted, where only the boundary and normal constraints are used. After introducing additional constraints into the framework, the outputs, depicted in the right column, are more consistent and do not contain artifacts
The first type of constraint is the negative normal constraint, which expands on the normal constraints to include the interior by displacing the boundary points one unit across the negative normal vector. This provides additional local information within the interior of the input to avoid artifacts near the boundary.
The second type of constraint is the grid constraint, which are points uniformly distributed about the current section. This grid layout of constraints captures global information and helps avoid having any “dead zones” in the function where the level of information may be low. The grid spacing can be adjusted to capture a finer grid along a section. To compute the \(h_i\) value for the grid constraints, a 3D Euclidean distance transform is applied to the input, and the distance values are retrieved at the constraint coordinates.
The third type of constraint is the propagated constraint. These constraints are propagated from adjacent sections, where their \(h_i\) values are recomputed for the current section, similar to the grid constraint. Propagated constraints work well for instances where the area of change between adjacent slices is significantly pronounced and can capture finer changes in local features but come with the downside of increasing computation significantly by carrying over constraints for nearly every section. As a result, propagated constraints are recommended in cases where the sections may differ markedly from the adjacent sections.
Different constraint types are depicted on the same left ventricular section. Each constraint is classified as a boundary (green), interior (cyan), or exterior (red) point, depending on its distance from the volume. Including these points adds local and global information to allow for more accurate interpolations and representations of input data
3.5.3 Application
When representing multiple structures within the heart, we can approach the implicit surface representation in one of two ways. First, we can represent all the structures within a volume in a single system. Implicit functions guarantee no self-intersection of surfaces of intermediary outputs due to constraints from each structure being factored into interpolation. However, computations in this approach can be time-intensive.
Alternatively, we can represent each structure separately as an individual implicit surface, which is faster and allows us to preserve label information and retain information about separate structures. Due to the arrangement of structures within the heart, the main structures are often separated by tissue or are otherwise relatively isolated. When we use implicit surfaces separately, there may be potential self-intersections between two or more different surfaces. In our case, the organ contours are extracted and are perfectly delineated in every time step. Because of smooth interpolation and imposition of additional constraints as explained in Sect. 3.5.2, individual organ contours do not intersect. An example can be seen in Fig. 6.
When representing structures as implicit surfaces, the computed weights are stored in memory. Interpolation operations can be performed by providing sets of coordinates (x, y, z, t) as input to the function, which outputs corresponding values that can be used to generate a discretized volume, as seen in Figs. 6, 8, and 9.
Output of an interpolation operation on an MRI exam is shown. Multiple structures of interest are present. The output on the left and its cross section on the right maintain a clean and smooth anatomy. The accuracy of these interpolated sections is shown in Table 3
3.5.4 Ejection fraction
Ejection fraction is a critical metric used to measure the ability of the heart to pump blood at each heartbeat interval. Computing this metric involves analyzing the left ventricle throughout the cardiac cycle in 4D MRI data. Ejection fraction can be calculated by dividing the difference between the left ventricular end-diastolic volume and the left ventricular end-systolic volume by the left ventricular end-diastolic volume, noted in Eq. (4). Estimating the ejection fraction can be done within our framework. We estimate ejection fraction with MRI data in the 2017 ACDC MICCAI challenge data set, where the end-diastolic (before contraction) and end-systolic (after contraction) phases have been delineated within the data.
To generalize our calculation, we find the volume metrics associated with the end-diastolic volume and the end-systolic volume within the left ventricle. This can be done by querying a grid of points into the implicit function and retrieving the outputs. Points mapped to a positive value are considered within the volume and are used for volume computation. We retrieve the minimum and maximum volume values from the implicit function and associate them with the end-systolic and end-diastolic volumes, respectively. Applying Eq. (4) gives us an estimated ejection fraction value between 0 and 100. This method has the advantage of making the computation of the volume and ejection fraction a straightforward and quick task.
4 Results
4.1 Segmentation
We reconstruct the patient’s heart using PyVista [46] and compare our segmentations to the ground truth annotations. Our single-threaded Python implementation on a 512 \(\times \) 512 \(\times \) 354 data set takes 10–15 min to segment—averaging 2 sec/image on an AMD Ryzen 9 5900X 12-Core Processor. A 3D visualization of the reconstruction of the heart model of a sample CT dataset is shown in Fig. 7 (Tables 1, 2).
For each image slice in the patient data, we compute the Sørensen–Dice coefficient, intersection over union (Jaccard), precision, recall, Hausdorff distance, and mean surface distance metrics between our binarized segmentation and ground truth for the following methods:
-
1.
Our method, using morphological geodesic active contours from distance map and watershed initializations with propagation.
-
2.
Three variants of our method:
-
(a)
Using only watershed initializations with propagation (ablation).
-
(b)
Using only distance map initializations with propagation (ablation).
-
(c)
Using active contours with no prior initialization and with propagation (ablation). All contour operations start from a circle of radius 5 on the centroids retrieved in Sect. 3.3.3.
-
(a)
-
3.
Using UNet 3+ [20] trained on the public MM-WHS 2017 CT training data set [53]. Training was set at 30 epochs using the Adam optimizer at a learning rate = 3e-4.
-
4.
Using only morphological geodesic active contours [34] to segment images. All contour operations start from a circle of radius 5 on the centroids from Sect. 3.3.3. There are no initialized segmentations from watershed/distance transform and no propagation.
-
5.
Using only watershed [35] to segment from centroids retrieved in Sect. 3.3.3.
-
6.
Using only thresholding [36] to segment (using values from Sect. 3.2.1).
4.2 Implicit surfaces
We demonstrate results from implicit surface interpolation for our data and compare the computation of the ejection fraction from our implicit representations.
4.2.1 Interpolation
Figures 8 and 9 depict interpolation operations between 2D and 3D keyframes, respectively.
An interpolation operation between two 2D slices of the left ventricle is depicted in the top row. The bottom row depicts an interpolation operation for multiple structures (myocardium, left ventricle, right ventricle, and pulmonary artery). Index 0 and 1 serve as keyframes, while index 0.5 is the discretized interpolated output
Depicted are the discretized output of a temporal interpolation operation between two 3D keyframes of the left ventricle. The volumes at index 0 and 1 serve as keyframes, while index 0.5 is the interpolated output. An outline of the previous frame is shaded in green to highlight the difference from the previous time step. The contraction at the green outlines and the mouth structure at the base of the LV indicate good interpolation results
To gauge interpolation accuracy, we interpolate various labeled 3D data sets and compare the original ground truth volumes in Table 3. We compare the interpolation output for 20 MRI exams from the MM-WHS data set [53], our annotated CT exam, 45 MRI exams from the Sunnybrook data set [38], and 50 pairs of diastolic and systolic phase instanced sections from the 2017 ACDC MICCAI challenge data set [4]. An example interpolation output from the MM-WHS data set is given in Fig. 6.
4.2.2 Ejection fraction
We estimate the ejection fraction from our left ventricular implicit representations as denoted in Sect. 3.5.4 and compare them to the ground truth values. All ejection fraction estimates were done on the ACDC MICCAI 2017 4D training data set [4], outputting ejection fraction values between 0 and 100. The absolute difference between our estimations and ground truth is depicted in Fig. 10. Our experiments show that our results closely approximated the ground truth, with a correlation coefficient of 0.983 and an average absolute difference of 2.728.
Ejection fraction is computed from the interpolation framework and compared with the ejection fraction of the ground truth exams. The red points indicate patient exams. The absolute differences between our estimated ejection fraction and ground truth ejection fraction values are depicted, with a 0.983 correlation coefficient
5 Discussion
From observing the results in Sect. 4, we evaluate the performance of our methods. Our pipeline can output consistent segmentations and achieves high accuracy with Dice scores and high robustness with precision scores among the metrics tested compared to other segmentation methods. Using morphological geodesic active contours, watershed, and thresholding by itself allows us to segment some slices but with high variance in scores. Initializing contours with segmentations and propagating these labels help to address these issues. Using watershed as an initializer for contouring garners a significant improvement and more consistent results than using watershed by itself; this however is still subject to watershed’s weaknesses, where some substructures will not be identified and contoured should watershed does not catch it in its initialization. This may be due to the noisy nature of cardiac image data, where boundaries can be more ambiguous, coupled with the watershed algorithm’s high susceptibility to noise. UNet 3+ produces better segmentation results than classical segmentation methods but has lower overall scores than our method; the quality and distribution of training data may be limited by the low amount of available data for whole-heart segmentation tasks (MM-WHS 2017 has a limited number of training exams). Our segmentation framework can provide sufficient segmentation results standalone; the nature of using initial segmentations as input also allows our framework to augment existing segmentation frameworks, such as using state-of-the-art CNN segmentation outputs as initial segmentation inputs to iterate on these outputs to cover deficiencies in image scanning technology or network segmentation output.
Our approach has multiple advantages: patient examinations are often noisy and important anatomical substructures often do not have clearly defined boundaries, making segmentation difficult. Morphological geodesic active contours are ideal for these segmentation problems, as they can detect boundaries through noisy and unclear data while maintaining intrinsic geometric measures of the image through the continued iterated evolution of contours. Using a balloon force, multiple contour initializations, and propagation from adjacent image sections allows for more robust segmentation and minimizes the amount of under-segmentation from watershed and active contours when encountering local minima. The automatic generation of initial contour segmentations also eliminates the need for user input. We additionally provide a robust framework for representing cardiac data in implicit form and allow for accurate and smooth interpolation across keyframes. From our results, our pipeline offers a robust semantic segmentation process and a promising implicit surfaces framework. All code will be made publicly available in a GitHub repository.
5.1 Future work
Our work has many potential applications and venues for future work in medical imaging. As we utilize contour growth approaches, future venues can incorporate using the output segmentations of convolutional neural networks-based approaches as input. During the imaging process, artifacts may arise from the scanner due to technical malfunction or patients not following breath-hold instructions. Similarly, cardiac arrhythmia, high heart rates, and scanner-EKG disconnections or malfunctions may impose challenges, resulting in suboptimal raw data. These artifacts may introduce noise that can significantly affect the final segmentation outputs of the scanner software. Limitations in software may then require manual correction which can be improved through an iterative contour growth approach without needing to re-scan or re-segment exams, using automatic generation or other segmentation outputs as priors. Additionally, abnormal segmentations between sections or frames due to scanner technical deficiencies and artifacts can be detected, and implicit functions can be fitted between keyframes to interpolate and fill in insufficient or distorted information.
A potentially promising application and extension of implicit surfaces in cardiology can involve synchronizing 4D patient data with EKG signals. Establishing correspondences between keyframes in 4D data to points within continuous 1D EKG signals can allow for dynamic visualization and representation of 3D intermediary heart states at any timestep of the EKG.
Data Availability
Our CT dataset is provided on figshare at https://figshare.com/s/caa70792c43ac01d8175. Public challenge data set is provided within the manuscript and manuscript references.
References
Appleton, B.: Optimal geodesic active contours: application to heart segmentation. In: APRS Workshop on Digital Image Computing (2003)
Bai, J.W., Li, P.A., Wang, K.H.: Automatic whole heart segmentation based on watershed and active contour model in ct images. In: 2016 5th International Conference on Computer Science and Network Technology (ICCSNT), pp. 741–744 (2016)
Balakrishnan, G., Zhao, A., Sabuncu, M.R., et al.: An unsupervised learning model for deformable medical image registration. CoRR arXiv:1802.02604 (2018)
Bernard, O., Lalande, A., Zotti, C., et al.: Deep learning techniques for automatic MRI cardiac multi-structures segmentation and diagnosis: Is the problem solved? IEEE Trans. Med. Imaging 37(11), 2514–2525 (2018)
Bui, I., Bhattacharya, A., Wong, S.H., et al.: Role of advanced three-dimensional visualization modalities in congenital heart surgery. Vessel Plus 6, 31 (2022)
Bui, V., Hsu, L.Y., Shanbhag, S.M., et al.: Improving multi-atlas cardiac structure segmentation of computed tomography angiography: a performance evaluation based on a heterogeneous dataset. Comput. Biol. Med. 125, 104019 (2020)
Caselles, V., Kimmel, R., Sapiro, G.: Geodesic active contours. Int. J. Comput. Vis. 22(1), 61–79 (1997)
Chan, T., Vese, L.: Active contours without edges. IEEE Trans. Image Process. 10(2), 266–277 (2001)
Chen, C., Qin, C., Qiu, H., et al.: Deep learning for cardiac image segmentation: a review. Front. Cardiovasc. Med. 7 (2020). https://doi.org/10.3389/fcvm.2020.00025
Chen, X., Williams, B.M., Vallabhaneni, S.R., et al.: Learning active contour models for medical image segmentation. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (CVPR) (2019)
Cohen, L.D.: On active contour models and balloons. CVGIP: Image Underst. 53(2), 211–218 (1991)
Dice, L.R.: Measures of the amount of ecologic association between species. Ecology 26(3), 297–302 (1945)
El-Taraboulsi, J., Cabrera, C.P., Roney, C., et al.: Deep neural network architectures for cardiac image segmentation. Artif. Intell. Life Sci. 4, 100083 (2023)
Fonseca, C.G., Backhaus, M., Bluemke, D.A., et al.: The cardiac atlas project-an imaging database for computational modeling and statistical atlases of the heart. Bioinformatics 27(16), 2288–2295 (2011)
Frangi, A., Niessen, W., Viergever, M.: Three-dimensional modeling for functional analysis of cardiac images: a review. IEEE Trans. Med. Imaging 20, 2–25 (2001)
Gharleghi, R., Chen, N., Sowmya, A., et al.: Towards automated coronary artery segmentation: a systematic review. Comput. Methods Programs Biomed. 225, 107015 (2022)
Habijan, M., Babin, D., Galić, I., et al.: Overview of the whole heart and heart chamber segmentation methods. Cardiovasc. Eng. Technol. 11(6), 725–747 (2020)
Habijan, M., Leventići, H., Galići, I., et al.: Whole heart segmentation from ct images using 3d u-net architecture. In: 2019 International Conference on Systems, Signals and Image Processing (IWSSIP), pp. 121–126 (2019)
Hemalatha, R., Thamizhvani, T., Dhivya, A.J.A., et al.: Active contour based segmentation techniques for medical image analysis. In: Koprowski, R. (ed.) Medical and Biological Image Analysis, chap. 2. IntechOpen, Rijeka (2018)
Huang, H., Lin, L., Tong, R., et al.: Unet 3+: a full-scale connected unet for medical image segmentation. In: ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP), pp. 1055–1059 (2020)
Iglesias, J.E., Sabuncu, M.R.: Multi-atlas segmentation of biomedical images: a survey. Med. Image Anal. 24(1), 205–219 (2015)
Kass, M., Witkin, A., Terzopoulos, D.: Snakes: active contour models. Int. J. Comput. Vis. 1(4), 321–331 (1988)
Kikinis, R., Pieper, S., Vosburgh, K.: 3D Slicer: a Platform for Subject-Specific Image Analysis, Visualization, and Clinical Support, vol. 3, pp. 277–289. Springer, New York (2014)
Klein, S., Staring, M., Murphy, K., et al.: elastix: a toolbox for intensity-based medical image registration. IEEE Trans. Med. Imaging 29, 196–205 (2010)
Krizhevsky, A., Sutskever, I., Hinton, G.E.: Imagenet classification with deep convolutional neural networks. In: Pereira, F., Burges, C., Bottou, L., Weinberger, K. (eds.) Advances in Neural Information Processing Systems, vol. 25. Curran Associates Inc. (2012)
Lederman, C., Joshi, A., Dinov, I., et al.: Tetrahedral mesh generation for medical images with multiple regions using active surfaces. Proc. IEEE Int. Symp. Biomed Imaging 2010, 436–439 (2010)
Lee, H.Y., Codella, N.C.F., Cham, M.D., et al.: Automatic left ventricle segmentation using iterative thresholding and an active contour model with adaptation on short-axis cardiac mri. IEEE Trans. Biomed. Eng. 57(4), 905–913 (2010)
Leow, L.J.H., Azam, A.B., Tan, H.Q., et al.: A convolutional neural network-based auto-segmentation pipeline for breast cancer imaging. Mathematics (2024). https://doi.org/10.3390/math12040616. https://www.mdpi.com/2227-7390/12/4/616
Liu, Z., Liu, F., Chen, W., et al.: Automatic segmentation of clinical target volumes for post-modified radical mastectomy radiotherapy using convolutional neural networks. Front. Oncol. (2021). https://doi.org/10.3389/fonc.2020.581347. https://www.frontiersin.org/journals/oncology/articles/10.3389/fonc.2020.581347
Lorensen, W.E., Cline, H.E.: Marching cubes: a high resolution 3d surface construction algorithm. SIGGRAPH Comput. Graph. 21(4), 163–169 (1987)
Menet, S., Saint-Marc, P., Medioni, G.: Active contour models: overview, implementation and applications. In: 1990 IEEE International Conference on Systems, Man, and Cybernetics Conference Proceedings, pp. 194–199 (1990)
Moe-Byrne, T., Evans, E., Benhebil, N., et al.: The effectiveness of video animations as information tools for patients and the general public: a systematic review. Front. Digt. Health 4, 1010779 (2022). https://doi.org/10.3389/fdgth.2022.1010779
Mumford, D., Shah, J.: Optimal approximations by piecewise smooth functions and associated variational problems. Commun. Pure Appl. Math. 42, 577–685 (1989)
Máirquez-Neila, P., Baumela, L., Alvarez, L.: A morphological approach to curvature-based evolution of curves and surfaces. IEEE Trans. Pattern Anal. Mach. Intell. 36(1), 2–17 (2014)
Najman, L., Schmitt, M.: Watershed of a continuous function. Signal Process. 38(1), 99–112 (1994). Mathematical Morphology and its Applications to Signal Processing
Nugroho, P.A., Basuki, D.K., Sigit, R.: 3d heart image reconstruction and visualization with marching cubes algorithm. In: 2016 International Conference on Knowledge Creation and Intelligent Computing (KCIC), pp. 35–41 (2016)
Pluempitiwiriyawej, C., Moura, J.M.F., Wu, Y.J.L., et al.: Stacs: new active contour scheme for cardiac mr image segmentation. IEEE Trans. Med. Imaging 24, 593–603 (2005)
Radau, P.E., Lu, Y., Connelly, K., et al.: Evaluation framework for algorithms segmenting short axis cardiac MRI. MIDAS J. (2009). https://doi.org/10.54294/g80ruo
Ranjan, A., Bolkart, T., Sanyal, S., et al.: Generating 3d faces using convolutional mesh autoencoders. In: Proceedings of the European Conference on Computer Vision (ECCV), pp. 704–720 (2018)
Ronneberger, O., Fischer, P., Brox, T.: U-net: convolutional networks for biomedical image segmentation. In: Navab, N., Hornegger, J., Wells, W.M., Frangi, A.F. (eds.) Medical Image Computing and Computer-Assisted Intervention—MICCAI 2015, pp. 234–241. Springer, Cham (2015)
Schreier, J., Genghi, A., Laaksonen, H., et al.: Clinical evaluation of a full-image deep segmentation algorithm for the male pelvis on cone-beam ct and ct. Radiother. Oncol. 145, 1–6 (2020). https://doi.org/10.1016/j.radonc.2019.11.021. https://www.sciencedirect.com/science/article/pii/S0167814019334917
Sharif, H., Rehman, F., Rida, A., et al.: A quick review on cardiac image segmentation. In: 2022 International Conference on IT and Industrial Technologies (ICIT), pp. 1–5 (2022)
Soomro, S., Akram, F., Munir, A., et al.: Segmentation of left and right ventricles in cardiac MRI using active contours. Comput. Math. Methods Med. 2017, 8350–680 (2017)
Sørensen, T.J.: A method of establishing group of equal amplitude in plant sociobiology based on similarity of species content and its application to analyses of the vegetation on danish commons. In: Biologiske Skrifter, vol. 5, pp. 1–34. Kongelige Danske Videnskabernes Selskab (1948)
Suinesiaputra, A., Medrano-Gracia, P., Cowan, B.R., et al.: Big heart data: advancing health informatics through data sharing in cardiovascular imaging. IEEE J. Biomed. Health Inform. 19(4), 1283–1290 (2015)
Sullivan, B., Kaszynski, A.: PyVista: 3D plotting and mesh analysis through a streamlined interface for the Visualization Toolkit (VTK). J. Open Source Softw. 4(37), 1450 (2019)
Tan, Q., Gao, L., Lai, Y.K., et al.: Mesh-based autoencoders for localized deformation component analysis. In: Proceedings of the AAAI Conference on Artificial Intelligence, vol. 32, p 1 (2018)
Teh, C.H., Chin, R.: On the detection of dominant points on digital curves. IEEE Trans. Pattern Anal. Mach. Intell. 11(8), 859–872 (1989)
Turk, G., O’Brien, J.F.: Shape transformation using variational implicit functions. In: Proceedings of the 26th Annual Conference on Computer Graphics and Interactive Techniques, SIGGRAPH ’99, p. 335–342. ACM Press/Addison-Wesley Publishing Co., USA (1999)
World Health Organization: Noncommunicable Diseases: Progress Monitor 2022. WHO Noncommunicable Diseases Progress Monitor Reports (2022)
Yeung, P.H., Namburete, A.I.L., Xie, W.: Sli2vol: Annotate a 3d volume from a single slice with self-supervised learning. In: 24th International Conference on Medical Image Computing and Computer Assisted Intervention–MICCAI 2021: Part II, pp. 69–79 (2021)
Zhang, Q., Eagleson, R., Peters, T.M.: Volume visualization: a technical overview with a focus on medical applications. J. Digit. Imaging 24(4), 640–664 (2011)
Zhuang, X., Shen, J.: Multi-scale patch and multi-modality atlases for whole heart segmentation of MRI. Med. Image Anal. 31, 77–87 (2016)
Acknowledgements
The authors would like to acknowledge Dr. Diana Glovaci from UCI Cardiology for her assistance with annotating our CT data.
Author information
Authors and Affiliations
Contributions
A.T. wrote the main manuscript text and prepared the figures. M. G. and H.S. provided feedback on the manuscript contents. Z.P. and I.P. provided resources for cardiology data and studies. All authors reviewed the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Supplementary file 1 (mp4 5760 KB)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Thai, A., Gradus-Pizlo, I., Pizlo, Z. et al. Automatic segmentation and implicit surface representation of dynamic cardiac data. Vis Comput (2024). https://doi.org/10.1007/s00371-024-03486-0
Accepted:
Published:
DOI: https://doi.org/10.1007/s00371-024-03486-0