Abstract
Visual analytics (VA) is a paradigm for insight generation by using visual analysis techniques and automated reasoning by transforming data into hypotheses and visualization to extract new insights. The insights are fed back into the data to enhance it until the desired insight is found. Many applications use this principle to provide meaningful mechanisms to assist decision-makers in achieving their goals. This process can be affected by various uncertainties that can interfere with the user decision-making process. Currently, there are no methodical description and handling tool to include uncertainty in VA systematically. We provide a unified workflow to transform the classic VA cycle into an uncertainty-aware visual analytics (UAVA) cycle consisting of five steps. To prove its usability, three real-world applications represent examples of the UAVA cycle implementation and the described workflow.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
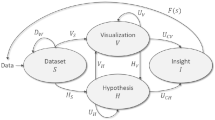
The increasing amount of data to be analyzed led to a novel data processing concept, defined as visual analytics (VA) [1]. Keim et al. [2] described VA as a connected process of four major components (Dataset S, Hypothesis H, Visualization V, and Insight I). These components are connected by functions that allow the transformation and analysis of the given input dataset while creating new insights. By now, VA is a discipline utilized in many applications to find novel insights in datasets, enabling effective decision-making [3].
Real-world applications and the resulting datasets used in VA are often affected by uncertainty originating from data incompleteness, unknown parameters, reconstruction artifacts, or the recognition process [4]. Hypothesis tests may use incomplete models and visualizations map data to color values, potentially suppressing information. In addition, the user introduces uncertainty through his cognitive abilities and personal biases. As a result, each component in the VA cycle can be affected by uncertainty, which needs to be quantified, propagated, and communicated throughout a VA cycle. Maack et al. [5] provided a general description of an uncertainty-aware visual analytics (UAVA) cycle, used as the basis for the workflow here.
They describe a VA cycle consisting of six components. The Dataset (S) is transformed into a UA Dataset (\(\overline{S}\)) that is processed by the UA Hypothesis and UA Visualization components to create UA Insight. This process is carefully monitored by the Provenance component to ensure the communication and understanding of uncertainty development throughout the cycle.
However, a general workflow to construct such a UAVA cycle and the process of including various sources of uncertainty is not available, as shown in Sect. 2. This forms the motivation of the presented work, starting with a brief recap of the basics of uncertainty that includes the definition and quantification of uncertainty, as shown in Sect. 3.
We provide a workflow that transforms a classic VA cycle, as provided by Keim et al. [2], into a UAVA cycle [5]. It covers the sources and effects of uncertainty in VA (see Sect. 4). The steps show the quantification of various types of uncertainty and to properly include them in a VA cycle. As a result, it constructs the UAVA cycle as defined in the work of Maack et al. [5].
We apply this workflow to three real-world applications: protein analysis, principal component analysis, and explainable artificial intelligence for predicting brain injury in Sect. 5. Each example presents an implementation of the proposed workflow. Finally, the approach will be discussed in Sect. 6.
In this work, we contribute:
-
A summary of workflows in the UAVA design space
-
A workflow to create UAVA applications
-
Hands-on examples of created UAVA
2 Related work
Keim and Zhang [6] stated that “Dealing with uncertainty in VA is nontrivial.” Based on this statement, Maack et al. [5] provided a general description of a UAVA cycle. Furthermore, Gillmann et al. [7] systematically described potential uncertainties in the VA cycle. Still, neither article yields a systematic approach to constructing such a cycle.
2.1 Uncertainty-aware visual analytics
UAVA approaches have been designed for multiple data types and computational models such as multivariate time series [8], principal component analysis [9], merge trees [10], or tensor analysis [11].
UAVA has tackled specific application scenarios approaches such as keyhole surgery assistance [12], analysis of Choropleths [13], and wildfire analysis [14]. Although these approaches show suitable UAVA approaches in specific target areas, they cannot be generalized to arbitrary use cases. Additionally, they lack a systematic method to include uncertainty in the VA cycle.
Sacha et al. [15] formulated the requirements to obtain a UA visualization. The requirements are built upon each other and suggest quantification, propagation, and interactive visualization of uncertainty in each component of the VA cycle. We will use these requirements to provide a workflow that leads to the UA description of the VA cycle defined by Maack et al. [5].
Correa et al. [16] provide a mathematical description of the requirements of Keim et al. [2]. Although they give hints on the construction of a UAVA cycle, a systematic description is not available.
Karami [17] provided a UAVA cycle that allows the processing of large datasets. Their work includes instantiated descriptions for big data of each component in the VA cycle. Similarly, this was accomplished for spatiotemporal data by Senaratne [18] to incorporate uncertainty. Although the problem of uncertainty in VA is quite known, a generalized workflow to create UAVA cycles does not exist, which forms the motivation for this work.
Uncertainty awareness is a problem that relates to many disciplines. These can be sensitivity analysis or VA of ensembles. These include ensemble visualization [19], sensitivity analysis [19], and data quality management [20]. These disciplines are highly related to UAVA and might serve as an inspiration for solutions. Still, a systematic workflow in these disciplines to create such visualization does not exist.
Summary: Various descriptions of UAVA and related issues exist. Still, the reviewed works miss a systematic workflow to create a UAVA approach.
2.2 Workflows and guidelines for uncertainty visualization
Haegele et al. [21] provided several examples of workflows for several applications such as hierarchical graphs and texts. Padilla et al. [22] summarized the potential design space for uncertainty visualization and provided existing visualization theories, which aim to minimize the large design space.
Frequency framing [23] is based on the idea that uncertainty is more intuitively understood in a frequency framing (e.g., 1 out of 10). Further, attribute substitution [24] allows viewers to mentally substitute uncertainty information for data that are easier to understand. Visual boundaries [25] separate a continuous space into a bounded space for easier recognition. At last, visual semiotics [26] provide visual variables suitable to indicate uncertainty. Although these are guidelines for visualizing uncertainty, they usually do not capture concepts such as provenance and interaction, which are important aspects of VA. This is a clear research gap detected for this work.
Summary: Many approaches aim to give guidelines for creating uncertainty-aware visualization. As VA is a broader topic, these cannot be applied immediately in the given context. Therefore, no workflow is available to design VA approaches.
2.3 Workflows to generate visual analytics
Gleicher et al. [27] defined the problem space to design visualizations. Their approach defines six axes along the problem space that must be considered while designing visualization. Although this gives a good starting point for visualization design, the approach does not cover the problem space of VA design. Booth et al. [28] provided a visual analytics space based on user goals without handling uncertainty. We will use this as a starting point in the presented work.
Conlen et al. [29] provided VA design principles that are mainly data-driven. They suggest how data should be visualized and which interactions should be provided. Unfortunately, these guidelines do not include uncertainty in their suggestions.
Federico et al. [30] provided a nested VA design model based on a user–task–data model. Here, the use of a VA system needs to be described to generate it.
There exist (open) software solutions that aim to guide users through the creation process of VA, such as RapidMiner, ParaView, and Tableau. They assist users in creating VA cycles. However, they do not provide systematic approaches to handling uncertainty.
Summary: We identified multiple works addressing workflows and guidelines for creating visualization or visual analytics approaches. Unfortunately, these do not holistically tackle uncertainty. Therefore, so far no workflow for creating UAVA is available.
UAVA cycle by Maack et al. [5]. The cycle consists of 6 major components that are connected
2.4 Research statement
Our presented work addresses three targeted aspects and formulates an overall research statement. The overall goal is to provide a workflow that can design UAVA applications, fusing and extending the knowledge of:
-
Uncertainty-aware visual analytics
-
Workflows and guidelines to create visualization solutions
-
Workflows and guidelines to design visual analytics
We recognize that this work is closely related to the work of Maack et al. [5]. However, here we present a significant improvement, as we do not focus on the definition of the UAVA space or cycle but on the description of design guidelines that allow the implementation of UAVA in applications. Specifically, Maack et al. describe the UAVA cycle and its connections but do not provide guidance on the creation of these cycles. This is a challenging task given the complexity of the UAVA cycle, and it raises questions about where to start, how to proceed, and which steps are necessary in what order.
3 Uncertainty-aware visual analytics
Maack et al. [5] defined an extension of the VA cycle (see Fig. 1) by Keim et al. [6], which will serve as a starting point and a goal in the presented workflow. This section is intended to summarize the presented cycle by Maack et al. to serve as a basis for the following considerations. The cycle consists of components that are connected by operations. Here, novel and adapted components and connections, in contrast to the classic VA cycle, are highlighted by a bar over their label.
3.1 Components of the uncertainty-aware visual analytics cycle
Six major components exist in the UAVA cycle.
Dataset S, is a very general concept that consists of n records \((r_1, r_2,..., r_n)\), where each record \(r_i\), consists of m observations, variables or attributes \((a_1, a_2,..a_m)\). An attribute \(a_i\) is a single entity, such as a number or symbol. A Dataset holds a structure that can be syntactic or semantic.
Uncertainty-aware Dataset \(\overline{S}\) is resulting from the input dataset S in conjunction with the extracted uncertainty quantification \({\overline{Q_S}}\). It describes the input dataset in conjunction with proper uncertainty quantification.
Hypothesis \(\overline{H}\) is a supposition or proposed explanation created based on limited evidence as a starting point for further investigation. To start, the null hypothesis is usually utilized. A hypothesis is formed and tested and can be either rejected or fail to be rejected. In conjunction with the null hypothesis, an uncertainty quantification is attached to it.
Visualization \(\overline{V}\) is an uncertainty-aware visual representation that the user can interpret.
Insight \(\overline{I}\) can be defined as knowledge gained during analysis and has to be internalized, synthesized, and related to prior knowledge including the uncertainty related to this insight.
Provenance \(\overline{P}\) When running a UAVA cycle, uncertainty will propagate and aggregate along with the operations carried out on the VA cycle. This implies the tracking of uncertainty throughout each computational step of the VA cycle, referred to as provenance.
3.2 Connections of the uncertainty-aware visual analytics cycle
The components of the UAVA cycle are related to each other using the following connections:
-
\(D_W: S \rightarrow S\), the preprocessing of a dataset
-
\(\overline{D_W}: \overline{S} \rightarrow \overline{S}\), the preprocessing of an uncertainty-aware dataset
-
\({\{\overline{Q_S}, \overline{Q_H}, \overline{Q_V}, \overline{Q_I}\}}: \{S, H, V, I\} \rightarrow \overline{S}, \overline{H}, \overline{V}, \overline{I}\), the uncertainty quantification for all components of the UAVA cycle
-
\(\overline{H_{\{S, V\}}}: \{\overline{S}, \overline{V}\} \rightarrow \overline{H} \), the generation of a UA hypothesis from a UA dataset \(\overline{S}\) or visualization \(\overline{V}\)
-
\(\overline{V_{\{S, H\}}}: \{\overline{S}, \overline{H}\} \rightarrow \overline{V} \), the generation of a UA visualization from dataset \(\overline{S}\) or hypothesis \(\overline{H}\)
-
\(\overline{U_{\{V, H\}}}: \{\overline{V}, \overline{H}\} \rightarrow \{\overline{V}, \overline{H}\} \), user interaction with the UA visualization \(\overline{V}\) or UA hypothesis \(\overline{H}\)
-
\(\overline{U_{\{CV, CH\}}}: \{\overline{V}, \overline{H}\} \rightarrow \overline{I} \), user interaction to generate UA insight \(\overline{I}\) from UA hypothesis \(\overline{H}\) or UA visualization \(\overline{V}\)
-
F(S), a feedback loop to insert generated insight I back into the VA cycle
-
\(\overline{F(S)}\), an uncertainty-aware feedback loop to insert generated uncertainty-aware insight \(\overline{I}\) back into the VA cycle
-
\(\overline{P_{S, H, V, I}}: {\overline{P}} \rightarrow \overline{P}\), the generation of Provenance from monitoring the state of \(\overline{S}, \overline{H}, \overline{V}, \overline{I}\)
These connections allow a transition between the components and guide through the analytic process.
4 Toward a systematic workflow
In the following, we propose a workflow comprising five steps to create a UAVA cycle, starting with a classic VA cycle. Various works exist that assist VA cycle design, and the resulting UAVA cycle can be constructed using most of the existing components and connections in the classic VA cycle.
4.1 Step 1: design a visual analytics cycle
The first step of the presented workflow is to create a classic VA approach (Fig. 2) that may be skipped when such an application is already present. It is highly dependent on the task, user, and dataset, as these categories imply the selection of proper hypothesis, visualization, and interaction techniques.
Keim and Zhang [6] show the general description of the classic VA cycle. This step selects all components and connections in the classic VA cycle necessary for the task. This may not include all connections that are present in the general description of the VA cycle.
In this stage, it is important to note that required data transformation (\(D_W\)) operations and the feedback loop (F(S)) are identified. Conlen et al. [29] provided a first attempt to provide design rules for VA approaches. These have been further refined and sorted into a workflow by Wang et al. [31]. We will use these stages as substeps in the presented workflow to generate a VA application.
Step 1.1 Content gathering and aggregation First, the content and information necessary for the task are collected by checking existing solutions and workflows. Users identify, seek, and extract information sources relevant to analytical tasks in their domain, meaning that they identify all data relevant to the task to create the Dataset component S.
Step 1.2 Content filtering and customization In this step, users use filtering to familiarize themselves with the collected content. They also personalize the analysis environment in which this content is filtered to fit their workflow. Here, the data preprocessing steps \(D_W\) are defined. This step is important, as data is often overwhelming, or not all aspects are necessary in an analysis scenario.
Step 1.3 Content organization and information analysis Users organize and examine the collected content from multiple perspectives to look for data patterns and desired information. Multiple levels of granularity can be found where different visualization techniques are needed. This refers to creating the visualization component in the VA cycle, where numerous visualization and interaction approaches must be examined, creating the connections \(V_S\) and \(U_V\).
Step 1.4 Evidence collection and hypothesis generation Possible automated tests and testable hypotheses are found next, collecting supporting evidence in the process. Sources of evidence for supporting or rejecting a hypothesis are identified, defining clear statements about the outcomes of a test. This is related to creating the Hypothesis component in the VA cycle. Proper analysis methodologies are established, resulting in the connections \(H_S\) and \(U_H\).
Step 1.5 Report generation and status update Connections between visual exploration and hypothesis testing are established. Visualization of analysis status and hypothesis tests via visual exploration is defined by providing notifications and updates processes. This creates the connections \(V_H\) and \(H_V\).
Step 1.6 Post-analysis and summarization Here, users focus on analysis outcomes of the implemented workflows, defining possible insight classes. Those insights are returned into the existing Dataset component, resulting in the feedback loop F(S).
At this point, uncertainty is not included in the design process explicitly. We acknowledge that the choices made in this step greatly impact the further design of a UAVA application. Still, this step comes with multiple benefits. First, it allows the systematic creation of the components and connections in the visual analytics cycle. Second, these components are also part of the UAVA cycle and help in the further design process as all potential sources of uncertainty are already defined.
4.2 Step 2: Quantify system uncertainty of the given visual analytics cycle
As shown in the work of Gillmann et al. [7], sources of uncertainty in VA can be separated into system and human uncertainties. Here, the focus is on the system uncertainties.
Based on the classic VA cycle in Step 1, system component uncertainty quantification can be performed, and the taxonomy of system uncertainties can be considered. The taxonomy provides a variety of information for each uncertainty event, namely uncertainty type, level of quantifiability, and a description.
For each source of uncertainty, the following steps need to be accomplished.
Step 2.1 Decide whether an uncertainty source affects the considered VA cycle Notably, not all sources of uncertainty may be inherent in every VA scenario. Further, there exist cases where sources of uncertainty may be present and known but are deemed irrelevant. This decision is made by the users, depending on the given task.
Step 2.2 Select types and levels of relevant uncertainty events According to the taxonomy of uncertainty events [7], the selected uncertainty events’ type and level can be collected.
Step 2.3 Determine the uncertainty description The uncertainty description of an event holds the following information: dimension, type, and continuity. This information depends on the data, hypothesis, and visualization the uncertainty event is attached to. Therefore, the description of the selected uncertainty events can be derived.
Step 2.4 Select a proper quantification strategy for the selected uncertainty events In the taxonomy of uncertainty events [7], each uncertainty event comes with one or more potential groups of uncertainty strategies. Together with the domain experts, available techniques are discussed and selected.
This creates three novel connections in the UAVA cycle: \({\overline{Q_S}}\), \({\overline{Q_H}}\) and \({\overline{Q_V}}\), as shown in Fig. 3. Importantly, quantifying uncertainty events in the component S (Dataset) leads to a novel \(\overline{S}\) component. In contrast to the other components, this separation is particularly important as not all uncertainty events are quantifiable with current knowledge. When applying a feedback loop, the incompleteness of quantifiability needs to be taken into account. This issue will be further investigated in Step 4.
4.3 Step 3: Integrate and connect uncertainty-aware solutions
This step combines the quantified uncertainty events and the selected methodologies. First, the selected methodologies from Step 1 are exchanged to achieve uncertainty-aware descriptions. For example, if a clustering approach is selected as the hypothesis, the approach needs to be exchanged with a UA clustering approach. This builds the UA components of the VA cycle, as shown in Fig. 4.
To implement the connections between the components in the VA cycle, uncertainty quantification approaches and the chosen UA methodologies need to be connected. Two operations achieve this: propagation of uncertainty and aggregation of uncertainty. Propagation is requested to connect the components in the VA cycle, whereas aggregation is required when multiple sources of uncertainty need to be summarized.
Please note that the components and connections in the VA cycle that have been revised to be UA are marked by a bar above their description in this manuscript (see Fig. 4). This results in the following three substeps.
Step 3.1 Exchange hypothesis H by an Uncertainty-aware hypothesis \(\overline{H}\) When selecting algorithms for exchange with uncertainty-aware variants, sensible decisions are made toward their improved effect on the workflow. By now, users can select from massive amounts of uncertainty-aware analysis tools. The first choice is recommended to be algorithms that are close to the originally selected algorithm while including uncertainty.
Step 3.2 Exchange visualization V by an uncertainty-aware visualization \(\overline{V}\) This step works simultaneously with Step 3.1. To find proper uncertainty-aware visualization techniques, state-of-the-art approaches shown in Sect. 2 can be considered.
Step 3.3 Aggregation of uncertainty in \(\overline{H}\) and \(\overline{V}\) As shown by Maack et al. [5], uncertainty can be introduced into the VA cycle in all components, where each component can be affected by multiple sources of uncertainty.
Technically, arbitrary aggregation functions can be used to achieve uncertainty aggregation, where Cai et al. [32] provided a survey of these functions. In the VA process, aggregation functions need to be selected so that all identified sources of uncertainty of one component can be aggregated properly. In addition, we suggest letting users select the importance of available uncertainty sources. This allows for creating awareness about the effect of different sources of uncertainty while rating them.
Step 3.4 Propagation of uncertainty between \(\overline{V}\) and \(\overline{H}\) When considering a transition from one component of the VA cycle into another, data needs to be transferred in a specific manner. This is highly dependent on the transformation step in the VA cycle.
Mathematical operations propagate data throughout the VA cycle (at least in the system component). These operations affect the data as well as their uncertainty. In addition, the inclusion of uncertainty also affects the operator, as they usually aim to map models known to be affected by uncertainty. As a result, operators need to be adjusted to handle data and their uncertainty. Multiple approaches, usually built on the principle of error propagation [33], exist to tackle this problem.
Step 3.5 Exchange insight I by an uncertainty-aware insight \(\overline{I}\)
The insights created by visual exploration or hypothesis testing are propagated into the insight component I together with attached uncertainty information. This uncertainty information can generally be directly propagated toward the insights, making the Insight component UA \(\overline{I}\). Also, the user could try to quantify the uncertainty about the extracted insight himself \(\overline{Q_I}\), i.e., by annotating the data or specifying his confidence as a number.
4.4 Step 4: Separate the visual analytics cycle feedback loop into quantifiable and unquantifiable feedback
The classical VA cycle has a feedback loop F(S) in which new insights are fed back into the Dataset component. This is the main advantage of the classic VA cycle, which aims to preserve and enhance the presented approach. Unfortunately, we showed that insight is hard to quantify, even partially, under the aspect of uncertainty. Here, the separation of new insights into two feedback loops is obtained to distinguish between both types, the classic feedback loop F(S) and the UA feedback loop \(\overline{F(S)}\), as shown in Fig. 5. The UA feedback loop can then be fed back into the UA Dataset component \(\overline{S}\), whereas the feedback loop that cannot be quantified with uncertainty will be fed back into the Dataset S component. As an example, this can be insights that arise from the process of exploring the shown dataset. Users might identify patterns in datasets that cannot be described mathematically. Here, the user must be incorporated to determine a proper uncertainty quantification. In this process, the identification of meaningful uncertainty quantification (\({\overline{Q_S}}\)) is extremely important.
Step 4.1 Define insight with uncertainty quantification \(\overline{S}\) Insights with uncertainty quantification need to be inserted in the \(\overline{S}\) component due to the missing need for uncertainty quantification. This insight usually results from the hypothesis component, which computes knowledge based on uncertainty-aware data and the uncertainty-aware hypothesis.
Step 4.2 Define insight without an uncertainty quantification S Insights without uncertainty quantification that need to be reinserted in the VA cycle must be fed back into the dataset component S. This justifies the existence of the original Dataset component S in the UAVA cycle, as not all gathered information can be quantified according to uncertainty. Starting from here, a suitable uncertainty quantification needs to be found according to the data structure of the insight. As the uncertainty of insight cannot be computed directly in many cases, insight can be modeled as a normal dataset and then be transferred into a UA Dataset through a suitable uncertainty quantification as described in Gillmann et al. [7].
4.5 Step 5: Generate provenance of uncertainty-aware components in the visual analytics cycle
When running a UAVA cycle, uncertainty will be propagated and aggregated along with the performed operations of the VA cycle according to the rules defined in Step 3. Varga et al. [34] highlighted the importance of provenance analysis and provided potential visualization approaches. This implies that uncertainty tracking is required throughout the entire VA cycle. This process is called provenance. As Maack et al. [5] proposed, a novel component in the VA cycle is required that assists in implementing provenance principles (P), as shown in Fig. 6.
As shown in the taxonomy of sources of uncertainty [7], these can be separated into sources of the system and human components. Provenance generation needs to be tackled differently in both cases.
Step 5.1 Provide provenance of sources of uncertainty in the system component On creation of a UA Dataset \(\overline{S}\), a UA Hypothesis \(\overline{H}\) or a UA Visualization \(\overline{V}\), the quantification of uncertainty, the operations utilized, and their results must be stored and made accessible.
Maack et al. [5] suggested providing visualization and interaction tools to let users follow uncertainty development throughout the VA process. This development aims to provide users with a tool to measure which operators cause a high increase in uncertainty. Additionally, thresholds that indicate whether the uncertainty exceeds a maximum, making the underlying data non-interpretable, should be available. While Herschel et al. [35] provided a survey on provenance creation, selecting a suitable tool or visualization approach highly depends on the underlying use case and the data type. In addition, Ragan et al. [36] characterized the provenance in visualization and data analysis approaches, which form the basis of the following discussion.
The provenance of a Dataset S focuses on the history of changes and movement of data. This type of provenance is very important in computational simulations and scientific visualization, as data is massively processed. The history of data and the respective uncertainty changes can include subsetting, data merging, formatting, transformations, or the execution of a simulation to ingest or generate new data [36]. These principles can be applied directly to \(\overline{S}\) and \(\overline{H}\) while creating two novel connections \({\overline{P_S}}\) and \({\overline{P_H}}\).
To achieve visualization and interaction provenance, Xu et al. [37] provided a survey summarizing existing technologies. We suggest checking for available techniques and carefully integrating them into the generated VA cycle, creating a novel connection \({\overline{P_V}}\).
Step 5.2 Provenance of sources of uncertainty in the human component
The provenance of UA Insights must also include uncertainty. Unlike data computations, insights are not directly observable as they are subjective, so their uncertainty is not observable directly as well [36]. A taxonomy of uncertainty sources for insights can be found in the work of Gillmann et al. [7].
This means that the provenance generation of the uncertainty sources contained in the Insight component can only be partially accomplished. Mechanisms exist that can be applied to tackle this problem.
As Ragan et al. [36] suggested, insight can be tracked by annotations. We suggest applying the same principle to the sources of uncertainty in the human component of the VA process. Here, two steps must be accomplished:
-
1.
Describe sources of uncertainty in the human component
-
2.
Provide an annotation tool to track occurring sources of uncertainty in the human component
4.6 Summary
Based on the discussion above, the resulting workflow to create UAVA assembles as follows:
-
1.
Design a visual analytics cycle
-
2.
Quantify system uncertainty of the given visual analytics cycle
-
3.
Integrate and connect uncertainty-aware solutions
-
4.
Separate the visual analytics cycle feedback loop into quantifiable and unquantifiable feedback
-
5.
Generate provenance of uncertainty-aware components in the visual analytics cycle
5 Example usage scenarios
To show the applicability of the presented workflow, we apply it to three different VA scenarios that originate from the scientific and information visualization areas.
We show how these approaches could have been built systematically based on the presented workflow. Please note that we will not discuss the visualization approach used and its benefits to the domain scientists in this section. Further details of the visualization and its benefits in the application domain can be found in the references of each use case.
5.1 Analysis of proteins
The UA protein analysis tool by Maack et al. [38] uses the positional uncertainty of atoms, provided by an anisotropic or isotropic model of their movement, to highlight regions of high disorder. The tool visualizes the uncertainty in a 3D visualization and a 2D Ramachandran plot, supplying the user with additional information in a detailed statistics view, as shown in Fig. 7. In the following, the design process is depicted along the five major steps, following the respective substeps in the process.
Design the VA application At first, researchers in this scenario work with PDB (Protein Data Bank) files to check the stability of new simulated data. Other tools like Chimera [39], and Polyview 3D [40], iMol [41], either become too cluttered, show only one instance of a protein at a time, or use too simplistic depictions for their analysis.
The data provides atom positions and secondary structure information (helix-shaped, sheet-shaped, or random coil), but the dihedral angles, i.e., the angle between the planes spanned by four neighboring backbone atoms using the middle atoms for the intersecting line, are not present yet and are calculated by the researchers in a preprocessing step. The combined information forms their Dataset component.
The researchers want to visualize their data using 3D renderings of the protein geometry and the 2D Ramachandran plot to control the stability of the protein. For 3D, the atom positions and secondary structure information are processed into various forms like ball-and-stick, solvent-accessible surface, or ribbon-style visualizations. The Ramachandran plot uses the preprocessed dihedral angles. Both visualization types fill the visualization component.
Most researchers’ queries revolve around the stability of molecules, as newly simulated variants often exhibit instabilities that lead to defects and unattainable designs. The Ramachandran plot and color mapping of the B-value are available in this scenario to test the stability, providing a visual hypothesis test for the Hypothesis component and connecting it to the visualization component.
When analyzing the data, the researchers find unstable regions that must be optimized as their Insight. Those regions are enhanced in further simulations to generate new designs to be tested, powering the feedback loop.
Quantify system uncertainty In designing the VA application, the researchers used the B-value, representing the isotropic movement of atoms, via color maps or analyzed multiple simulation results of the same protein by superposition. Still, they want to understand better the extent and effect of the B-value or spread of the ensemble of molecules.
They find that the B-value partially stems from finite instrument resolution providing a fixed standard deviation per atom, and the ensemble resembles variations in observations due to minor changes in the starting conditions, providing an anisotropic distribution. Both types of uncertainty are 3D and quantifiable, allowing them to be represented in the 3D visualization. Those properties also hold true for the respective dihedral angles in each scenario (isotropic and anisotropic). Also, non-representative sampling could be a problem but is discarded as the software is not meant to integrate simulation software at this point. Mapping, interpolation, and shading are minor sources of uncertainty and are therefore disregarded in this application. It is known that they are schematic representations of atoms and proteins, so the deviations in their exact form or slight variations of the color coding do not strongly influence the decision-making.
Both selected types of uncertainty are Level 2, making them fully quantifiable. They are 3D or 2D with respect to the atom position or dihedral angle distribution. The B-value gives a discrete standard deviation, whereas the eigendecomposition defines a continuous ellipsoid of deviation around each atom or angle combination.
As the B-value directly corresponds to the standard deviation of movement per atom, uncertainty quantification is present via a given formula and propagated straight to the computation of dihedral angles. Eigenvalues and vectors must be computed via eigendecomposition for the ensemble of simulated molecules. Similarly, the distribution of the dihedral angles in the Ramachandran plot is decomposed into eigenvalues and vectors per amino acid residue.
Integrate and connect uncertainty-aware solutions
When testing the stability of a molecule, the researchers need to find statistics to support or reject their hypothesis. In addition to standard descriptions of atoms, their uncertainty in each eigenvector direction is provided here. Alternatively, the standard deviation extracted from the B-value can be shown.
To ensure accurate data visualization, it is essential that the Visualization component can effectively process the uncertainty information provided by the Data component. This crucial step will help ensure that the insights gleaned from the visualizations are reliable and can be used to make informed decisions. The researchers want to understand how far the atoms can move under the underlying uncertainty models. As blurring seemed to clutter the visualization too much in other applications, they want a contour around their visualizations that can be set to a user-defined threshold in standard deviations. Therefore, a scalar field is created that describes the distance of each point in space to the respective protein geometry in directional standard deviations. The researchers can interactively change this distance using the marching tetrahedra/triangles algorithm with different thresholds. The procedure is similar for both 3D and 2D visualizations in this application.
As the researchers are interested in finding amino acid residues (the building blocks of molecules) that make the protein unstable, the uncertainty of individual atoms can be summarized and displayed per amino acid residue.
Also, individual amino acid residues can be marked by the researchers to connect hypothesis testing with the visualization, delivering details on demand.
Feedback loop separation Defining the VA cycle’s feedback loop, the user’s interaction with the application must be considered. As shown in Fig. 7, the UA visualizations of the protein and the Ramachandran plot are shown next to each other, providing a statistics view with numerical data. When a user selects an atom in the 3D visualization, the Ramachandran plot highlights the corresponding dihedral angle pair and vice versa. Also, numerical information about the angles and considered atoms is shown, including their uncertainty information. The impressions obtained by the user cannot be directly quantified at this stage. Still, the uncertainty information from UA Insights can be used first to improve the regions most affected by uncertainty. This way, the most problematic configurations are handled first, still giving the user more regions to look at and ordering them by uncertainty. This feedback can be used to generate new simulations with other software solutions that can be analyzed again in the present application.
Provenance generation Implementing Provenance is carried out by the possibility to view all protein models in a single visualization while providing the distribution of models, showing them as iso-surfaces around the average model. To keep track of the individual models, the researchers can interactively scroll through them while maintaining uncertainty information, allowing them to select strong outliers. Here, Provenance uncertainty is only possible when using anisotropic models. In the isotropic case, the uncertainty directly stems from the uncertainty of the average model with the eigenvectors attached, created by summarizing all models.
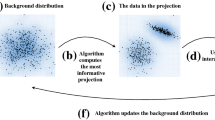
UA PCA by Görtler et al. [42]. The PCA is computed based on probability distributions and covariance matrices that form the uncertainty quantification. The influence of uncertainty can be followed by a user-adjusted value in the smaller views on the left providing a mechanism of provenance
Insight provenance could be recorded by the average uncertainty of the dataset in each cycle iteration. It would be helpful to implement such a procedure to integrate simulations into the present application, tracking the uncertainty in each step. This would allow users to find occurrences where changes to certain areas resulted in higher overall uncertainty levels. The tracked uncertainty levels could then be made explorable by a variety of provenance exploration tools.
5.2 Uncertainty-aware principal component analysis
Görtler et al. [42] presented a VA approach for UA principal component analysis (PCA), as shown in Fig. 8. The proposed method holds an extended PCA approach and a VA framework to explore the results of the analysis. We chose this example as PCA is an important algorithm often considered in the context of data analysis.
As input data, PCA requires high-dimensional data in the first step. Here, any high-dimensional data that is numerical can be considered (Step 1.1). Depending on the given task, the important axes are selected. In Step 1.3, the two-dimensional space is chosen, whereas the PCA technique is determined in Step 1.4. At last, the extracted dimension reduction can be fed back into the visual analytics cycle. As a result, we obtain a classic visual analytics cycle by the definition of Keim et al.
To quantify uncertainty in each component, the method considers a statistical model. This maps to the second step in the workflow. Here, the uncertainty in the data component is described as random points and vectors. The uncertainty in the component hypothesis is described using uncertain covariance matrices, an extension of classic covariance matrices. The selected source of uncertainty is of level 2. The event is described in the same dimensionality as the considered points of the high-dimensional dataset. As we can describe this uncertainty with random points this maps to the group of sensitivity analysis.
For step three, the hypothesis is exchanged by the UA extension of PCA. Further, to exchange the visualization, not solely the mapped points in the dataset are shown, but also their probability distribution. In addition, users are enabled to manipulate the covariance matrix’s influence factor in the performed PCA. This is also demonstrated as a plot (Step 3.4). Step 3.3 can be skipped in this context as only one source of uncertainty is considered.
The feedback loop is separated to implement step four. The resulting UA PCA mapping applies to feedback that can be quantified by uncertainty. For insight that cannot be quantified by uncertainty, proper selection of scaling factors and insights while manipulating it can be named.
For step five, the provenance is achieved directly by manipulating the scaling factor. Here, the direct consequences of the impact of uncertainty on the resulting PCA can be monitored while following the trace of the eigenvalues in the additional view.
5.3 Explainable artificial intelligence for brain lesion prediction
Our last example refers to the ability of VA to allow users to understand the models used in AI. In the presented example of Gillmann et al. [43], a neural network is used to predict the lesion in a patient’s brain that can result from stroke, as shown in Fig. 9. The authors trained several networks all being able to predict the lesion to a certain accuracy, but the medical collaborators wished to understand how the network made its decision [44].
VA for analyzing the neural network to predict a brain lesion by Gillmann et al. [43]. Left: slice view to compare ground truth and prediction. Upper right: histogram of the prediction. Lower right: ROC of made prediction
In the first step, we start with the content gathering (Step 1.1). Here, data related to the stroke imaging process can be considered. In the given case, the clinic used computed tomography scans enriched with contrast bolus to highlight the brain’s vascular system and perfused tissue (Step 1.2). The visual system was developed to compare the ground truth used for training to the neural network’s prediction, as shown in Fig. 9 (Step 1.3). As usual in such scenarios, the predicted lesion is thresholded by 0.5, creating our hypothesis (Step 1.4). Further, the display represents the histogram of the currently reviewed prediction and the ROC (receiver operator curve) of the made prediction. This information can be fed back into the visual analytics cycle (Step 1.5).
In the second step, all relevant sources of uncertainty must be quantified. In this specific case, there is an inherent uncertainty in the input dataset’s ground truth, as three ground truths exist per patient. This results in a level 2 uncertainty event with a dimensionality of 1. Additionally, the machine learning model introduces uncertainty into the VA process, as it is potentially incomplete. This can be expressed by a level 3 uncertainty event using sensitivity analysis (Step 2.1–2.4)
In the third step, the components of the VA cycle are exchanged. At first, the U-Net, which was used to predict the patient’s lesion, is exchanged by a Bayesian U-Net (Step 3.1). Further, the evaluation of the U-Net was adapted by an uncertainty-aware computation, as shown in Fig. 10 by Sperling et al. [45] (Step 3.2). Here, the hard threshold that decides which pixels are considered true and false positives, as well as true and false negatives, is exchanged by a probability distribution function. Adjusting \(\sigma \) of this threshold leads to areas in the classification that cannot be clearly sorted into the four classes.
UAVA evaluation of neural networks by Sperling et al. [45]. TP (green), FP (red), TN (blue), and FN (purple) are shown together. From left to right by manipulating the \(\sigma \) of the used uncertainty-aware threshold, separating the four classes becomes fuzzier (white areas)
The feedback loop must be separated in the fourth step. In this case, computations such as accuracy or other metrics, considering the quality of the prediction, can be computed based on the uncertainty-aware classification of pixels from the prediction (Step 4.1).
In the fifth step, the implementation of provenance is required. This directly relates to the manipulation of \(\sigma \), as users can directly relate the consequences of the used \(\sigma \).
6 Discussion
Building upon the taxonomy of sources of uncertainty in VA [7] and the UAVA cycle of Maack et al. [5], this paper describes a workflow that transforms a classic VA cycle into a UAVA cycle in five steps. The taxonomy adds components and connections to the VA cycle to quantify, propagate, and communicate the captured sources of uncertainty. This is done so that we can provide a mechanism that identifies and tackles uncertainty in the system component and monitors uncertainties in the human component.
Examples are provided for each step of the workflow, including detailed information on how a step manipulates the given VA cycle. The applicability of the presented workflow is shown by mapping it to three real-world application scenarios where uncertainty needs to be incorporated into the VA cycle. Here, two examples from scientific visualization and one from the information visualization area are presented. The examples span a wide range of data sources, indicating generality. Further applications, such as tumor detection or lesion prediction, have been tested but not included in this study. This shows that the presented workflow is capable of handling various scenarios.
Although we showed how to extend the known VA cycle to create uncertainty awareness, further research is necessary. Creating a novel UAVA cycle needs to be investigated in terms of potential applications and data sources. Our approach shows a clear strength in scientific visualization applications. Further applications originating from the information visualization domain, such as graph analysis or multi-dimensional data, need to be considered.
At last, the provided workflow is intended to give guidelines. Although we described each step in great detail, it leaves space for design choices. As design choices in a VA system are the key to success or failure, a working UAVA cycle cannot be guaranteed. Here, further research on specific VA approaches including their use cases and scenarios regarding uncertainty is required.
7 Conclusion and future work
We proposed a workflow that can transform a classic VA cycle into a UAVA cycle. Here, five steps need to be accomplished. First, a VA cycle is designed. Second, all sources of uncertainty must be detected and, if possible, quantified. Third, components in the VA cycle need to be exchanged for uncertainty-aware alternatives. Fourth, the feedback loop needs to be separated, and last, provenance mechanisms need to be implemented. For each step, we provide suggestions and list possibilities for accomplishing a successful implementation. Three real-world UAVA scenarios provide a post hoc analysis of these approaches, mapping them to the workflow. The protein analysis example provides a full design pipeline to also show possible decisions during the execution of the workflow. These examples show the applicability of the proposed workflow.
We aim to provide a state-of-the-art analysis of available UAVA approaches in future work. We also want to describe and define the boundary between ensemble visualization and uncertainty visualization approaches.
Data availability
Data sharing is not applicable to this article as no datasets were generated or analyzed during the current study.
References
Cui,W.: Visual analytics: A comprehensive overview, IEEE Access (2019)
Keim,D.A., Mansmann,F., Schneidewind,J., Thomas,J., Ziegler,H.: Visual analytics: scope and challenges. In: Visual data mining (2008)
Kohlhammer, J., May, T., Hoffmann, M.: Visual analytics for the strategic decision making process. In: GeoSpatial Visual Analytics (2009)
Griethe, H., Schumann, H. et al.: The visualization of uncertain data: methods and problems. In: SimVis (2006)
Maack, R.G.C., Scheuermann, G., Hernandez, J., Gillmann, C.: Uncertainty-aware visual analytics: scope, opportunities, and challenges. Vis. Comput. (2022). https://doi.org/10.1007/s00371-022-02733-6
Keim, D., Zhang, L.: Solving problems with visual analytics: challenges and applications. In: Proceedings of the 11th International Conference on Knowledge Management and Knowledge Technologies (2011)
Gillmann, C., Maack, R.G.C., Raith, F., Pérez, J.F., Scheuermann, G.: A taxonomy of uncertainty events in visual analytics. IEEE Comput. Gr. Appl. 43, 62 (2023)
Bors, C., Bernard, J., Bögl, M., Gschwandtner, T., Kohlhammer, J., Miksch, S.: Quantifying uncertainty in multivariate time series pre-processing. In: EuroVis Workshop on Visual Analytics (2019)
Görtler, J., Spinner, T., Streeb, D., Weiskopf, D., Deussen, O.: Uncertainty-aware principal component analysis. IEEE Trans. Vis. Comput. Gr. 26, 822 (2020)
Yan, L., Wang, Y., Munch, E., Gasparovic, E., Wang, B.: A structural average of labeled merge trees for uncertainty visualization. IEEE Trans. Vis. Comput. Gr. 26, 832 (2019)
Gerrits, T., Rössl, C., Theisel, H.: Towards glyphs for uncertain symmetric second-order tensors. Comput. Gra. Forum. 38, 325 (2019)
Gillmann, C., Maack, R.G.C., Post, T., Wischgoll, T., Hagen, H.: An uncertainty-aware workflow for keyhole surgery planning using hierarchical image semantics. In: Proceedings of PacificVAST 2018 (2018)
Huang, Z., Lu, Y., Mack, E., Chen, W., Maciejewski, R.: Exploring the sensitivity of choropleths under attribute uncertainty. IEEE Trans. Vis. Comput. Gr. 26, 2576 (2019)
Preston, A., Gomov, M., Ma, K.: Uncertainty-aware visualization for analyzing heterogeneous wildfire detections. IEEE Comput. Gr. Appl. 39, 72 (2019)
Sacha, D., Senaratne, H., Kwon, B.C., Ellis, G., Keim, D.A.: The role of uncertainty, awareness, and trust in visual analytics. IEEE Trans. Vis. Comput. Gr. 22, 240 (2016)
Correa, C.D., Chan, Y., Ma, K.: A framework for uncertainty-aware visual analytics. In: 2009 IEEE Symposium on Visual Analytics Science and Technology (2009)
Karami, A.: A framework for uncertainty-aware visual analytics in big data. In: CEUR Workshop Proceedings (2015)
Senaratne, H.V.: Uncertainty-aware visual analytics for spatio-temporal data exploration, Ph.D. thesis, Universität Konstanz (2017)
Wang, J., Hazarika, S., Li, C., Shen, H.-W.: Visualization and visual analysis of ensemble data: a survey. IEEE Trans. Vis. Comput. Gr. 25, 2853 (2018)
Liu, S., Andrienko, G., Wu, Y., Cao, N., Jiang, L., Shi, C., Wang, Y.-S., Hong, S.: Steering data quality with visual analytics: the complexity challenge. Vis. Inf. 2, 191 (2018)
Hägele, D., Schulz, C., Beschle, C., Booth, H., Butt, M., Barth, A., Deussen, O., Weiskopf, D.: Uncertainty visualization: fundamentals and recent developments. it Inf. Technol. 64(4–5), 121–132 (2022). https://doi.org/10.1515/itit-2022-0033
Padilla, L., Kay, M., Hullman, J.: Uncertainty visualization. In: Piegorsch, W.W., Levine, R.A., Zhang, H.H., Lee, T.C.M. (eds.) Computational Statistics in Data Science. Wiley, Oxford (2022)
Gigerenzer, G.: The psychology of good judgment: frequency formats and simple algorithms. Med. Decis. Mak. 16(3), 273–280 (1996). https://doi.org/10.1177/0272989X9601600312
Kahneman, D., Frederick, S.: Representativeness revisited: attribute substitution in intuitive judgment. In: Gilovich, T., Griffin, D., Kahneman, D. (eds.) Heuristics & Biases: The Psychology of Intuitive Judgment. Cambridge University Press, New York (2002)
Padilla, L.M., Creem-Regehr, S.H., Hegarty, M., Stefanucci, J.K.: Decision making with visualizations: a cognitive framework across disciplines. Cogn. Res. Princip. Impl. 3, 29 (2018). https://doi.org/10.1186/s41235-018-0120-9
MacEachren, A.M., Roth, R.E., O’Brien, J., Li, B., Swingley, D., Gahegan, M.: Visual semiotics & uncertainty visualization: an empirical study. IEEE Trans. Vis. Comput. Gr. 18(12), 2496–2505 (2012). https://doi.org/10.1109/TVCG.2012.279
Gleicher, M., Riveiro, M., von Landesberger, T., Deussen, O., Chang, R., Gillman, C.: A problem space for designing visualizations. IEEE Comput. Gr. Appl. 43(4), 111–120 (2023)
Booth, P., Gibbins, N., Galanis, S.: Design spaces in visual analytics based on goals: analytical behaviour, exploratory investigation, information design & perceptual tasks. In: Hawaii International Conference on System Sciences (2019)
Conlen, M., Stalla, S., Jin, C., Hendrie, M., Mushkin, H., Lombeyda, S., Davidoff, S.: Towards design principles for visual analytics in operations contexts. In: Proceedings of the 2018 CHI Conference on Human Factors in Computing Systems, CHI ’18, Association for Computing Machinery, New York, NY, USA, 2018, 1–7. https://doi.org/10.1145/3173574.3173712
Federico, P., Amor-Amorós, A., Miksch, S.: A nested workflow model for visual analytics design and validation. In: Proceedings of the Sixth Workshop on Beyond Time and Errors on Novel Evaluation Methods for Visualization, BELIV ’16, Association for Computing Machinery, New York, NY, USA, 2016, p. 104-111. https://doi.org/10.1145/2993901.2993915
Wang, X., Butkiewicez, T., Dou, W., Bier, E.A., Ribarsky, W.: Designing knowledge-assisted visual analytics systems for organizational environments (2011)
Cai, S., Gallina, B., Nyström, D., Seceleanu, C.: Data aggregation processes: a survey, a taxonomy, and design guidelines. Computing 101, 1397 (2018)
Ghilani, C.: Statistics and adjustments explained part 3: Error propagation, Lecture Notes (2004)
Varga, M., Varga, C.: Visual analytics: data, analytical and reasoning provenance. In: Building Trust in Information (2016)
Herschel, M., Diestelkämper, R., Ben Lahmar, H.: A survey on provenance: What for? what form? what from? VLDB J. 26, 881 (2017)
Ragan, E.D., Endert, A., Sanyal, J., Chen, J.: Characterizing provenance in visualization and data analysis: an organizational framework of provenance types and purposes. IEEE Trans. Vis. Comput. Gr. 22, 31 (2015)
Xu, K., Ottley, A., Walchshofer, C., Streit, M., Chang, R., Wenskovitch, J.: Survey on the analysis of user interactions and visualization provenance. Comput. Gr. Forum 39, 757 (2020)
Maack, R.G.C., Raymer, M.L., Wischgoll, T., Hagen, H., Gillmann, C.: A framework for uncertainty-aware visual analytics of proteins. Comput. Graph. 98, 293 (2021)
Pettersen, E.F., Goddard, T.D., Huang, C.C., Meng, E.C., Couch, G.S., Croll, T.I., Morris, J.H., Ferrin, T.E., ChimeraX, U.C.S.F.: Structure visualization for researchers, educators, and developers. Protein Sci. 30(1), 70–82 (2021). https://doi.org/10.1002/pro.3943
Porollo, A., Meller, J.: Versatile annotation and publication quality visualization of protein complexes using POLYVIEW-3D. BMC Bioinf. 8(1), 316 (2007). https://doi.org/10.1186/1471-2105-8-316
Piotr,R.: iMol Overview (2007). https://www.pirx.com/iMol/overview.shtml
Gortler, J., Spinner, T., Streeb, D., Weiskopf, D., Deussen, O.: Uncertainty-aware principal component analysis. IEEE Trans. Vis. Comput. Gr. (2020). https://doi.org/10.1109/tvcg.2019.2934812
Gillmann, C., Peter, L., Schmidt, C., Saur, D., Scheuermann, G.: Visualizing multimodal deep learning for lesion prediction. IEEE Comput. Gr. Appl. 41(5), 90–98 (2021). https://doi.org/10.1109/MCG.2021.3099881
Welle, F., Stoll, K., Gillmann, C., Henkelmann, J., Prasse, G., Kaiser, D.P., Kellner, E., Reisert, M., Schneider, H.R., Klingbeil, J. et al.: Tissue outcome prediction in patients with proximal vessel occlusion and mechanical thrombectomy using logistic models, Transl. Stroke Res. pp. 1–11 (2023)
Sperling, L., Lämmer, S., Hagen, H., Scheuermann, G., Gillmann, C.: Uncertainty-aware evaluation of machine learning performance in binary classification tasks (2022)
Acknowledgements
The authors acknowledge the financial support by the Federal Ministry of Education and Research of Germany and by the Sächsische Staatsministerium für Wissenschaft Kultur und Tourismus in the program Center of Excellence for AI-research “Center for Scalable Data Analytics and Artificial Intelligence Dresden/Leipzig,” project identification number: ScaDS.AI.
Funding
Open Access funding enabled and organized by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors have no conflict of interest to declare.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Maack, R.G.C., Raith, F., Pérez, J.F. et al. A workflow to systematically design uncertainty-aware visual analytics applications. Vis Comput (2024). https://doi.org/10.1007/s00371-024-03435-x
Accepted:
Published:
DOI: https://doi.org/10.1007/s00371-024-03435-x