Abstract
Dynamic tomography reconstructs a time activity curve (TAC) for every voxel assuming that the algebraic form of the function is known a priori. The algebraic form derived from the analysis of compartmental models depends nonlinearly on the nonnegative parameters to be determined. Direct methods apply fitting in every iteration step. Because of the iterative nature of the maximum likelihood–expectation maximization (ML–EM) reconstruction, the fitting result of the previous step can serve as a good starting point in the current step; thus, after the first iteration we have a guess that is not far from the solution, which allows the use of gradient-based local optimization methods. However, finding good initial guesses for the first ML–EM iteration is a critical problem since gradient-based local optimization algorithms do not guarantee convergence to the global optimum if they are started at an inappropriate location. This paper examines the robust solution of the fitting problem both in the initial phase and during the ML–EM iteration. This solution is implemented on GPUs and is built into the 4D reconstruction module of the TeraTomo software.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
1 Introduction
Dynamic positron emission tomography (PET) examines the dynamics of radiotracer accumulation in tissues [17, 19, 25]. The determined TAC of voxel V is searched in a predefined algebraic form \({\mathcal K}({\theta }_V, t)\) of time t and parameter vector \({\theta }_V\). The algebraic form is defined, for example, by compartmental models [5, 8, 28], i.e., as the solution of the differential equation of radiotracer exchange between compartments [29, 31].
In direct methods the kinetic model is integrated into the reconstruction algorithm. The measurement time is partitioned into intervals \((t_F, t_{F+1})\) called frames. The expected number of decays \({\tilde{x}}_F\) in frame F is
where \(\lambda \) defines the decay rate of the radiotracer. The maximum likelihood estimator finds voxel parameters \({\theta }_V\) that maximize the likelihood of the measured events. At the location of the extremum, the derivatives of the likelihood are zero, which leads to the following nonlinear equations for every voxel V and parameter P:
where \(x_{V,F}\) is the activity estimate of voxel V in frame F after a pair of static forward and back projections, which couples the equations of all voxels. For a computationally straightforward implementation, the nested EM algorithm [14, 25, 27] decouples the equations of voxels by using \(x_{V,F}\) from the previous iteration step. This method iterates two steps: The first executes forward and back projections in each frame to get updated voxel activity \(x_{V,F}\), and the second fits parameters \({\theta }_V\) in each voxel V. The significant benefit of the nested EM algorithm is that the fitting step can be executed independently for each voxel. In this case, the objective functions of the local fitting establish a surrogate of the likelihood, which is a Kullback–Leibler-like term:
Instead of maximizing the global likelihood, this error term is minimized independently in every voxel V. The gradient of this term is zero at the solution of Eq. (2). As a similar problem is solved in every pixel, from now on, voxel subscript V is removed from the formulas.
Having selected the fitting criterion, algorithms for finding the best fit are needed. The Levenberg–Marquardt algorithm [15] is often used for least-square fitting, especially when there are no constraints. However, Eq. (2) is not equivalent to least-square fitting.
The main difference is that the least-square fitting would minimize the absolute error between the data points and the fitted function, while Eq. (2) can be interpreted as the minimization of the relative error. Such problems can be attacked by replacing the \(1/{\tilde{x}}_F({\theta })\) term in Eq. (2) by its Taylor’s expansion [22]:
where \({\theta }^*\) is the current estimate and \(\mathbf {d} = {\theta } - {\theta }^*\) is the step to the new estimate. With this substitution, we get a linear system of equations for the unknown step
where the elements of matrix \(\mathbf{F}\) and vector \(\mathbf{d}\) are evaluated at current estimate \({\theta }^*\):
The organization of this paper is as follows. Section 2 reviews the related previous work, identifies the possibilities of improvement and presents the research objectives. Section 3 discusses the original and the modified algebraic descriptions of compartmental models. In Sect. 4, we present our analytic computation scheme for activity values and their derivatives. Section 5 discusses our model-specific improvement of the Levenberg–Marquardt scheme, and Sect. 6 proposes the method for finding appropriate initial values for the iterative optimization. Sections 7 and 8 compare the proposed methods with the state of the art in 2D reconstruction of a mathematical phantom and in the reconstruction of 3D data. The paper closes with conclusions.
2 Previous work, problem statement and objectives
The state of the art of dynamic positron emission tomography is reviewed in survey papers [4, 19, 25, 26]. Our paper belongs to the category of nonlinear fitting since the compartmental models are essentially nonlinear [29, 31]. Such fitting methods have several difficulties. The measured data are contaminated by noise [23, 24]. The actual guess of the kinetic model should be integrated in time and then derivated with respect to the optimization parameters. The optimization parameters are nonnegative and upper bounded. The error term of Eq. 3 is not quadratic, i.e., its derivative is not linear. Thus, this is a nonlinear, constrained fitting, which needs numerical methods as there is no direct analytic solution. Integrals can be estimated with numerical quadrature [30], but they have high computational cost and high error if the function changes quickly. Applied general numerical optimization techniques include coordinate descent [8], Newton–Raphson iteration [20], preconditioned gradient descent [16] and Levenberg–Marquardt iteration [28], which is most popular choice because it needs only the first derivatives and is simple to implement. Gradient-based optimization methods converge quickly close to the solution, but do not guarantee convergence, and even if they converge, they can be stuck in a local optimum. Sophisticated global optimization techniques would have high computational cost since this fitting should be executed for every voxel and in each ML–EM iteration [11, 12]. For gradient-based solvers, the specification of the initial state becomes crucial. Starting with constant initial values or initializing the parameters with random numbers may cause slow initial convergence. To attack this problem, we propose an initialization scheme that exploits the original measured data and is simple to compute; thus, its additional computational cost is amortized by the faster convergence.
Note that general optimization techniques do not take into account the particular properties of the PET problem. Problem-specific solutions include simplifications, e.g., the replacement of the nonlinear fitting by a much simpler least-square fitting [18]. Note that a direct solution would be available for the least-square fitting of a function that linearly depends on its parameters. However, modifying the optimization criteria would alter and slow down convergence. If only a subset of parameters have such linear behavior, the fitting can be decomposed to a nonlinear fitting and then the least-square fitting of the linear parameters [2, 7].
The main research goal of this paper is to propose algorithms that solve the fitting problem efficiently, i.e., more robustly and more accurately than the classical Levenberg–Marquardt method without significantly increasing the computation time. In particular, the main contributions of this paper are as follows:
-
A modification of the algebraic form of the two-tissue model is proposed in Sect. 3, which is exploited by the refinement of the Levenberg–Marquardt scheme in Sect. 5. Unlike previous work, our simplified fitting schemes do not replace the complicated but accurate nonlinear solvers, but provide additional potential refinements. Their improvement is checked, and if they fail to improve the fitting, their proposal is ignored. In our GPU implementation, the additional improvement trial has negligible additional computational cost.
-
We present the analytic computation of matrix \(\mathbf {F}\) and vector \(\mathbf {r}\) of the linear system of Eq. (5) as well as its automatic derivation in Sect. 4. The analytic computation not only leads to a straightforward implementation, but is also significantly faster than the numeric evaluation.
-
A low computational cost initial estimation process is established to guess the parameters from where the ML–EM iteration can be started (Sect. 6).
Our goal is a method that is applicable in clinical and preclinical practice and can solve reconstruction problems involving about a billion lines of responses and voxels in reasonable time, and therefore is appropriate for parallel GPU implementation. As GPUs prefer single-precision arithmetics, the proposed techniques are to be stable and robust to lower-precision computations.
3 Algebraic forms of compartmental models
Based on the solution of differential equations expressing the tracer exchange between n compartments [5], the TAC has an algebraic form that is a nonnegative linear combination of convolutions of the blood input function \(C_p(t)\) and the impulse response w(t) of the tissue. The impulse response is the non-negatively weighted sum of exponentials of unknown, nonnegative rate constants \(\alpha _i\) and constrained only for positive time values t by the Heaviside step function \(\epsilon (t)\):
where \(c_i\) factors are the nonnegative weights. Taking into account that a voxel is a mixture of the tissue and blood, we obtain
where \(f_v \in [0,1]\) is the unknown fraction of blood and \(C_W\) is the known or separately measured total blood concentration function taking also into account that portion of the radiotracer that cannot diffuse into the tissues.
Reconstruction means the identification of parameters \({\theta }=(f_v, c_1, \alpha _1, c_2, \alpha _2,\ldots )\) for every voxel. However, these parameters are not optimal for fitting since the TAC depends on these parameters nonlinearly. Thus, we use an equivalent algebraic form, where \(f_v, a_1, a_2, \ldots \) are linear parameters whenever the rate constants are fixed:
where the correspondence between the new and the original parameters is \(a_i = {(1-f_v)c_i}\).
Blood input functions \(C_p(t)\) and \(C_W(t)\) are also subjects to fitting executed once at the beginning of the reconstruction. Feng’s model [3] assumes them in the following algebraic form if the radiotracer is injected in \(t=0\):
if \(t>0\) and zero otherwise. Blood exponents showing the blood activity dynamics \(\beta _1, \ldots , \beta _4\) and linear blood parameters \(A_1, \ldots , A_4\) are determined with simulated annealing.
4 Analytic integration and derivation of the activity
Voxel activity requires the integration of the convolutions of exponential functions. During the iterative fitting of the kinetic model, the derivatives of the convolutions are needed. This section presents an analytic method with an easy program implementation to obtain these integrals and their derivatives.
According to Eq. (1), the activity of a voxel in a frame is expressed by a time integral. The antiderivative of the integrand is
where there are two types of convolutions of terms from the blood input function and the impulse response. Using the \(\alpha _i^* = \alpha _i+\lambda \) and \(\beta _j^* = \beta _j+\lambda \) notations, the first type is:
The second type of convolutions is
Finally, the activity in a frame is
During the fitting process, we also need the derivatives of these integrals with respect to the kinetic parameters:
where the derivatives of time integrals \(G_{ij}(t)\) and \(H_i(t)\) can still be analytically expressed, but become quite complicated.
Another problem is that the above formulas are invalid if \(\alpha ^*_i = \beta _j^*\) or \(\alpha ^*_i=0\) or \(\beta ^*_j=0\), and are numerically unstable when the values are close to these conditions. As \(\beta ^*_j\) blood exponents represent the dynamics of the blood activity, while rate constants \(\alpha ^*_i\) represent the dynamics of the tissue activity, it can easily happen that during the numerical optimization these values get very close to each other. On the other hand, irreversibility can also cause \(\alpha _i\) to go to zero.
Both the algebraic complexity of the derivatives and the need for special cases can be attacked by the application of dual numbers [1] in the computer implementation. This means that a function f is represented by a dual number \(f + {\mathbf {i}}f'\) where \(\mathbf {i}\) is the imaginary unit with the \({\mathbf {i}}^2=0\) definition, the real part is the value of function f and the imaginary part is the value of the derivative at the same location \(f'\). It is easy to see that the arithmetic rules of basic operations of addition, subtraction, multiplication and division for such dual numbers are the same as the original arithmetic rules in the real part and as the derivation rules in the imaginary part.
When we encounter a 0/0 type undefined division or the numerator and denominator are close to zero causing numerical instability, the l’Hospital rule can automatically be applied. If \(f_1(x)\) and \(f_2(x)\) are close to zero, then
Thus, we also need the second derivatives. To cope with this or with Newton–Raphson iteration, the dual numbers should be generalized for at least two imaginary units \(f + {\mathbf {i}}f' + {\mathbf {j}}f''\). The arithmetic rules of the imaginary units that make the basic operations similar to derivation rules are
The chain rule on the exponential has the following form:
This works well when convolution \(G_{ij}\) of Eq. (11) is computed. However, convolution \(H_i\) of Eq. (12) has \((\alpha ^*_i - \beta ^*_1)^2\) in its denominator, i.e., its derivative has \((\alpha ^*_i - \beta ^*_1)^4\) in its denominator. Thus, l’Hospital rule should be applied four times to obtain a nonzero value in the denominator, making the computation of derivatives up to the second order insufficient. It would be possible to further extend dual numbers, but the performance penalty would be too high. Therefore, when \(\alpha _i\) is very close to \(\beta _1\), it is perturbed and the derivative is computed a little farther.
Convolutions \(H_1\) and \(G_{11}\) plotted as functions of rate constant \(\alpha ^*_1\), setting \(\beta ^*_1 = 0.5\), \(t_F = 0\), \(t_{F+1} = 6\). Note that these functions have singularities at \(\alpha ^*_1 = 0\) and \(\alpha ^*_1 = \beta ^*_1 = 0.5\). The upper figure shows the results of analytic integration and automatic derivation. The lower figure depicts the numerical results when integrals are computed with step \(\varDelta t = 0.02\) and derivatives are computed with \(\varDelta \alpha = 10^{-5}\). We used single-precision arithmetic (float) in all cases
The arithmetic rules of the dual numbers can be summarized by a simple C++ class exploiting operator overloading. With this, we can implement only the computation of the integrated values, while the derivatives and the 0/0 type divisions are automatically taken care of. Figure 1 shows the \(H_1\) and \(G_{11}\) plotted as functions of \(\alpha ^*_1\) and compares our analytic approach to numerical integration and differentiation. The analytic approach is not only more accurate and robust, but is an order of magnitude faster to compute.
5 Fitting during iterative reconstruction
The solution of Eq. (2) can be interpreted as a nonlinear constraint fitting that minimizes the relative error. However, the simple approach of Euclidean norm and linear models can at least partially be used for more complex models. On the one hand, in Sect. 3 an equivalent algebraic form was proposed, in which a larger linear subgroup of the parameters can be identified. A set of parameters is said to form a linear subgroup if the model depends on them linearly when all parameters outside this group are fixed. On the other hand, we find a weighting scheme for the Euclidean norm, which can be minimized with a direct method. With these tricks, we can easily refine the parameter values of the linear subgroup. If the extra step of the simple fitting for the linear parameters does not improve the error term of Eq. 3, then the result of this extra step can be ignored.
Having obtained an estimate for the parameters by general constraint fitting, we can further refine the linear parameters \(\bar{{\theta }} = (f_v,a_1,a_2, \ldots )\) of activity \({\tilde{x}}_F\):
where vector \(\mathbf {b}_F^T=(b^{(0)}_F, b^{(1)}_F, b^{(2)}_F, \ldots )\) contains the integrals of the basis functions in frame F:
Note that these can be obtained analytically using the results of Sect. 4.
Fitting requires the solution of Eq. (2), which can be rewritten as:
As this phase starts with the estimates obtained with a nonlinear equation solver and then refines the linear parameters, we already have a guess \({\hat{x}}_F\) for \({\tilde{x}}_F({\theta })\) showing up in the denominator. Let us substitute \({\tilde{x}}_F({\theta })\) by \(\mathbf {b}_F^T \cdot \bar{\theta }\) in the numerator and the partial derivative:
Considering all linear parameters and rearranging the terms, a system of linear equations is obtained for the linear parameters:
This method is called the weighted refinement. If \({\hat{x}}_F\) is set to 1, which corresponds to a non-weighted least-square fitting, then the method is called linear refinement.
Solving the linear system of Eq. (21) may result in values that are outside of the allowed range, i.e., weights \(a_i\) may be negative and \(f_v\) outside of [0, 1]. Negative parameters could be removed by nonnegative least-square fitting, other violations by inequality constrained least squares. Here, we use a simpler technique. If some parameters are outside of the allowed range, their value is substituted by the boundary value that is overstepped and \(x_F\) is reduced by the product of the boundary value and the corresponding basis function. For the parameters inside the range, another linear system is constructed in the form of Eq. (21), but the elements of the fixed parameters are removed from basis vector \(\mathbf {b}\) reducing the size of the linear system to the number of not-fixed parameters. This operation is repeated until all parameters are inside the allowed range or on its boundary.
6 Initial estimation
The initial estimation of the parameters can use just the measured data, which is the list of events that are binned in frames, and thus can be expressed by a matrix \(y_{L,F}\) or a vector of LOR values \(\mathbf {y}_F\) in each frame F.
To find a guess for the voxel activities, we use a direct method and not iterative techniques, because at this stage, we do not have information about the order of magnitude of the activity values. Starting an iteration from a many order of magnitude different value may cause numerical instability in the GPU favouring 32 bit precision arithmetics.
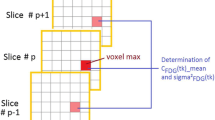
The initial direct estimation assumes that the volume can be partitioned into \(N_R\) homogeneous regions \({\mathcal R}_1, {\mathcal R}_2, \ldots , {\mathcal R}_{N_R}\) where all voxels belonging to the same region R share the same parameters and thus have the same activity values \({\tilde{x}}_{R,F}\). Thus, in this phase, the dimension of the problem is reduced from the number of voxels to the number of regions, simplifying the expected number of coincidences \({\tilde{y}}_{L,F}\) in LOR L during frame F to
where \({\mathbf{B}}_{L,R}\) is the region-based system matrix, storing the probabilities that a decay in region R causes a coincidence event in LOR L. Its row R can be obtained by forward projecting a volume of constant 1 values in voxels belonging to region R and zero elsewhere.
Reconstructions of macroparameters \(K_1\), \(V_D\) and \(K_i\) of the 120k coincidence measurement. The horizontal coordinate is the ground truth, and the vertical is the reconstructed value. Perfect reconstructions would be on the diagonal line. Purple dots depict white matter data and green dots gray matter data. The average errors are also shown below the plots
Reconstructions of macroparameters \(K_1\), \(V_D\) and \(K_i\) of the 12k coincidence measurement. The horizontal coordinate is the ground truth, and the vertical is the reconstructed value. Perfect reconstructions would be on the diagonal line. Purple dots depict white matter data and green dots gray matter data. The average errors are also shown below the plots
Because of considering regions rather than individual voxels, the Poisson model can be replaced by a Gaussian model since a region has much more decays than single voxels, and the Gaussian assumption is acceptable for high statistic measurements. Note that in the extreme case, we can consider the whole field of view as a single region. In case of the Gaussian model, the reconstruction is equivalent to the solution of the following linear system:
where \(y_{L,F}\) is the measured number of coincidences in LOR L during frame F. Comparing this equation to Eq. (22), we note that the expected number of the coincidences \({\tilde{y}}_{L,F}\) is replaced by the measured number of coincidences \(y_{L,F}\), which is a maximum likelihood estimation in case of Gaussian distribution. This overdetermined linear equation can be solved using the Moore–Penrose pseudo-inverse:
Executing this step for every frame, we have a discrete time activity function for every region. The next step is to obtain the initial parameters. Thanks to the down-sampling from voxels to a significantly smaller number of regions, sophisticated global optimization methods can be applied at this point. Rate constants \(\alpha _i\) are the essentially nonlinear part of the concentration function. If rate constants are known, the concentration depends on the other parameters linearly, which could be determined at least approximately with the solution of a linear system. For the initial estimation of the rate constants, we use either a grid of the possible values or explore this space with simulated annealing. When we take a point in this space, the rate constants are given a coordinates and other linear parameters are obtained by the discussed method for the linear subgroup, and the Kullback–Leibler-like error term (Eq. (3)) is evaluated and assigned to the point of rate constants. The goal of visiting grid points or simulated annealing is to find that point of rate constant where this fitting error is minimal.
So far, the results are based on the Gaussian assumption, which is justified by working with larger regions and not individual voxels. The initial estimates can be further refined by considering the Poisson model. It means that a few region-based ML–EM iterations are executed. As the number of regions is significantly less than the number of voxels, the complete initial guess has negligible computation time with respect to the reconstruction.
7 Results of 2D reconstruction
The proposed methods are tested first with a 2D phantom where the number of LORs is \(N_{L}=10\)k and the number of voxels is \(N_V=1024\) (Fig. 2). The 2D phantom has four anatomic parts, namely the air, gray matter, white matter and the blood. Parameters of the two-tissue compartment model \([f_v, a_1, a_2, \alpha _1, \alpha _2]\) used in the test are listed in Table 1 as reference values. Figure 6 depicts the reference blood input function and the TAC in different tissues. The total number of coincidences during the 10 second long measurement time is 120k. The measurement time is decomposed to 20 frames. The average number of coincidences per LOR per frame is about 0.6; thus, it would be a low statistic data for static reconstruction. In order to have an even lower statistic measurement, we also investigate the case of 12k coincidences. In this 2D test, we applied the method of sieves as a regularization, i.e., executed an anatomy-aware smoothing after each iteration step.
7.1 Initial guess of parameters
The initial guess assigns initial parameters to regions, so such regions should be defined. Now we use the anatomic parts as regions. Thus, to get the first guess of the time activity functions, just a linear system of Eq. (24) needs to be solved in each frame. The initial parameter fitting generates \(10^4\) samples in the 2D domain of the two rate constants, and the remaining linear parameters are found by solving a linear system of Eq. (21) for each sample. Figure 3 shows the error maps of different regions, where the color of a point depicts the error of the best fit when the two rate constants are defined by the coordinates of the point. This figure demonstrates that the search space is indeed complicated and has many local minima and maxima; thus, a robust global optimization method is necessary to initialize the rate constants. Then, to move toward the Poisson model, 20 region-based ML–EM iterations are executed.
Table 1 lists the resulting parameters after the global optimization and the region-based ML–EM iterations involving weighted refinement. Note that even these are fairly accurate since the test case meets the assumption that regions are uniform.
In the second round of tests, we used the described region-based initial parameter guessing method for sets of reference \(K_1, k_2, k_3, k_4\) kinetic parameters, which are generated randomly in the [0.02, 2] interval. With the random reference parameters, we have simulated 100 measurements of total coincidences between 14k and 900k. Reconstructed macroparameters \(K_1\), \(V_D\) and \(K_i\) are paired with the ground truth values and depicted as points in 2D plots of Fig. 4. This figure compares three options including also the average errors of the reconstructed parameters. In case of “ML–EM iterations,” the proposed initial guess is executed assuming a single region that includes all voxels, and then, 20 ML–EM iterations are executed with strong anatomy-aware regularization. The results of “Initial guess” are obtained executing the region-based initial parameter estimation. Finally, the method of “Initial guess + ML–EM iterations” refines the results of the initial estimation with 20 region-based ML–EM iterations. Let us observe the macroparameter error reduction due to the initial estimation and the added region-based ML–EM iteration. The same test has also been executed having reduced the activity to its tenth. The results from which similar conclusions can be drawn are shown in Fig. 5.
7.2 Fitting during the iterative reconstruction, without region-based initialization
In these tests, we investigate the effect of the refinement of linear parameters. The initial estimation is turned off, i.e., the volume is handled as a single region during parameter initialization. The \(K_1\) and \(V_d\) parametric images as well as the mean and the standard derivation of time activity functions are obtained after 50 iterations. In a single ML–EM iteration, two Levenberg–Marquardt sub-iterations are executed, which may be followed by the proposed linear or weighted refinement step. The added computation cost of the refinement is less than ten percent of a single Levenberg–Marquardt sub-iteration. Figure 6 shows TACs with reconstructed region activity average and standard deviation, and demonstrates the effect of the refinement of the linear parameters on the reconstructed time activity curves and macroparameter maps. The relative RMS error of the TAC curve of the Levenberg–Marquardt method is reduced from 13.9% to 9.1% and to 8.4% by the added linear refinement and weighted refinement, respectively. The difference is especially noticeable in the [0,1] second interval of the gray matter and white matter time activity functions in Fig. 6.
The reconstruction errors as functions of the number of iteration steps are depicted in Figs. 7 and 8, demonstrating that the refinement is worth executing as it further reduces the error. The weighted refinement is only slightly better than the linear refinement and is worth using in case of low statistic measurements.
8 Dynamic 3D reconstruction
The performance of the proposed method in 3D dynamic reconstruction is demonstrated with input data obtained by simulating the measurement of the Zubal phantom [33] by GATE [6] assuming the Mediso NanoScan PET/CT scanner. The “measured data” are reconstructed with our implemented system at \(128 \times 128 \times 64\) voxel resolution and compared to the ground truth. The 10 second long measurement time is partitioned into 20 frames of lengths inversely proportional to the activity. The brain phantom has nine different homogeneous regions, including gray matter, white matter, cerebellum, caudate nucleus, putamen, bone, skin, blood and air. In these tests, 20 full ML–EM iterations are executed, while we applied total variation (TV) regularization of the same parameter in all cases. Our method is orthogonal to the applied spatial regularization, so other regularization approaches like Bregman iteration [21], anisotropic diffusion [23] or sparse representation-based techniques could also be incorporated [9, 10, 32].
The proposed method including optional initial estimation and weighted refinement in iterations is compared to nonparametric reconstruction when frames are reconstructed independently and to the original nested ML–EM method. In our initial estimation, four major regions are distinguished: air, bone + skin, cerebellum and the composition of everything else. The TACs are shown in Fig. 9, and the reconstructed activities in frame 5 in Fig. 10. Direct, i.e., parametric reconstructions are superior to indirect, i.e., static reconstruction in all cases. The proposed initial estimation and refinement are also beneficial and improves the reconstruction with respect to the original nested ML–EM scheme. Note, for example, the more accurate TAC of the putamen and especially the caudate nucleus, and also the sharper boundary of the gray matter.
The execution times of different steps on an NVIDIA GeForce RTX 2080 GPU are shown in Table 2. The initial estimation time counts only once, the fitting time should be multiplied by the number of iterations, forward and back projection times by the number of frames and the number of iterations. The time of refinement is about 6% of the time of fitting. A full iteration with 20 frames takes 60 seconds, and the results are obtained by 20 iterations; thus, the 5 second long time required by the initial estimation is negligible.
9 Conclusions
This paper proposed improvements to the nested ML–EM algorithm to robustly and efficiently fit compartment models to noisy data. Both the initial estimation and the steps of the iterative solution have been addressed. We also discussed an analytic approach to compute the parameters of the Levenberg–Marquardt solver, which is not only precise but also much faster than the numerical estimation of the integrals of convolutions. During the iterative solution, a refinement step is included which proposes a modified value after the Levenberg–Marquardt solution, which may or may not be accepted using the local fitness criterion. All steps of the method are appropriate for GPU implementation. We have shown results obtained with a 2D phantom and also 3D reconstructions of GATE simulated data. This solution is integrated into the TeraTomo system [13].
References
Baydin, A.G., Pearlmutter, B., Radul, A.A., Siskind, J.: Automatic differentiation in machine learning: a survey. J. Mach. Learn. Res. 18, 1–43 (2018)
Cheng, X., Li, Z., Liu, Z., Navab, N., Huang, S.C., Keller, U., Ziegler, S.I., Shi, K.: Direct parametric image reconstruction in reduced parameter space for rapid multi-tracer pet imaging. IEEE Tran. Med. Imaging 34(7), 1498–1512 (2015)
Feng, D., Huang, S.-C., Wang, X.: Models for computer simulation studies of input functions for tracer kinetic modeling with positron emission tomography. Int. J. Bio-Med. Comput. 32(2), 95–110 (1993)
Gallezot, J., Lu, Y., Naganawa, M., Carson, R.E.: Parametric imaging with pet and spect. IEEE Trans. Radiat. Plasma Med. Sci. 4(1), 1–23 (2020)
Gunn, R.N., Gunn, S.R., Cunningham, V.J.: Positron emission tomography compartmental models. J. Cereb. Blood Flow Metab. 21(6), 635–652 (2001). PMID: 11488533
Jan, S., et al.: GATE: A simulation toolkit for PET and SPECT. Phys. Med. Biol. 49(19), 4543–4561 (2004)
Kadrmas, D.J., Oktay, M.B.: Generalized separable parameter space techniques for fitting 1k–5k serial compartment models. Med. Phys. 40(7), 072502 (2013)
Kamasak, M.E., Bouman, C.A., Morris, E.D., Sauer, K.: Direct reconstruction of kinetic parameter images from dynamic pet data. IEEE Trans. Med. Imaging 24(5), 636–650 (2005)
Li, X., Shen, H., Zhang, L., Zhang, H., Yuan, Q., Yang, G.: Recovering quantitative remote sensing products contaminated by thick clouds and shadows using multitemporal dictionary learning. IEEE Trans. Geosci. Remote Sens. 52(11), 7086–7098 (2014)
Liao, H. Y., Sapiro, G.: Sparse representations for limited data tomography. In: 2008 5th IEEE International Symposium on Biomedical Imaging: From Nano to Macro, pp. 1375–1378, (2008)
Liao, W., Lange, K., Bergsneider, M., Huang, S.: Optimal design in dynamic pet data acquisition: a new approach using simulated annealing and component-wise metropolis updating. IEEE Trans. Nucl. Sci. 49(5), 2291–2296 (2002)
Liu, X., Hou, F., Hao, A., Qin, H.: A parallelized 4d reconstruction algorithm for vascular structures and motions based on energy optimization. Vis. Comput. 31(11), 1431–1446 (2015)
Magdics, M., et al.: TeraTomo project: a fully 3D GPU based reconstruction code for exploiting the imaging capability of the NanoPET/CT system. In: World Molecular Imaging Congress, (2010)
Matthews, J. C., Angelis, G. I., Kotasidis, F. A., Markiewicz, P. J., Reader, A. J.: Direct reconstruction of parametric images using any spatiotemporal 4d image based model and maximum likelihood expectation maximisation. In: IEEE Nuclear Science Symposuim Medical Imaging Conference, pp. 2435–2441 (2010)
Moré, J.J.: The levenberg-marquardt algorithm: implementation and theory. In: Watson, G.A. (ed.) Lecture Notes on Computer Science, Springer, vol. 630, pp. 105–116 (1978)
Rahmim, A., Tang, J., Mohy-ud Din, H.: Direct 4d parametric imaging in dynamic myocardial perfusion pet. Front. Biomed. Technol. 1(1), 4–13 (2014)
Rahmim, A., Tang, J., Zaidi, H.: Four-dimensional (4d) image reconstruction strategies in dynamic pet: beyond conventional independent frame reconstruction. Med. Phys. 36(8), 3654–70 (2009)
Reader, A.J., Matthews, J.C., Sureau, F.C., Comtat, C., Trebossen, R., Buvat, I.: Iterative kinetic parameter estimation within fully 4d pet image reconstruction. In: IEEE Nuclear Science Symposium Conference Record vol. 3, pp. 1752–1756 (2006)
Reader, A.J., Verhaeghe, J.: 4d image reconstruction for emission tomography. Phys. Med. Biol. 59(22), R371 (2014)
Reutter, B.W., Sang Oh, Gullberg, G.T., Huesman, R.H.: Improved quantitation of dynamic spect via fully 4-d joint estimation of compartmental models and blood input function directly from projections. In: IEEE Nuclear Science Symposium Conference Record, vol. 4, pp. 2337–2341, (2005)
Szirmay-Kalos, L., Tóth, B., Jakab, G.: Efficient Bregman iteration in fully 3d PET. In: IEEE Nuclear science symposium and medical imaging conference, MIC’14, (2014)
Szirmay-Kalos, L., Kacsó, Á., Magdics, M., Tóth, B.: Dynamic pet reconstruction on the GPU. Period. Polytech. Electr. Eng. Comput. Sci. 62(4), 134–143 (2018)
Szirmay-Kalos, L., Magdics, M., Tóth, B.: Volume enhancement with externally controlled anisotropic diffusion. Vis. Comput. 33, 331–342 (2017)
Wang, C., Yang, J.: Poisson noise removal of images on graphs using tight wavelet frames. Vis. Comput. 34(10), 1357–1369 (2018)
Wang, G., Qi, J.: Direct estimation of kinetic parametric images for dynamic pet. Theranostics 3(10), 802–815 (2013)
Wang, G., Rahmim, A., Gunn, R.N.: Pet parametric imaging: Past, present, and future. IEEE Trans. Radiat. Plasma Med. Sci. 4(6), 663–675 (2020)
Wang, G., Qi, J.: Acceleration of the direct reconstruction of linear parametric images using nested algorithms. Phys. Med. Biol. 55(5), 1505–1517 (2010)
Wang, G., Qi, J.: An optimization transfer algorithm for nonlinear parametric image reconstruction from dynamic pet data. IEEE Trans. Med. Imaging 31(10), 1977–1988 (2012)
Watabe, H., Ikoma, Y., Kimura, Y., Naganawa, M., Shidahara, M.: Pet kinetic analysis–compartmental model. Ann. Nucl. Med. 20(9), 583 (2006)
Yan, J., Planeta-Wilson, B., Carson, R.E.: Direct 4-d pet list mode parametric reconstruction with a novel em algorithm. IEEE Trans. Med. Imaging 31(12), 2213–2223 (2012)
Yan, J., Planeta-Wilson, B., and Carson, R.E.: Direct 4d list mode parametric reconstruction for pet with a novel em algorithm. In: IEEE Nuclear Science Symposium Conference Record, NSS ’08, pp. 3625–3628, (2008)
Ye, J., Wang, H., Yang, W.: Image reconstruction for electrical capacitance tomography based on sparse representation. IEEE Trans. Instrum. Meas. 64(1), 89–102 (2015)
Zubal, I.G., Harrell, C.R., Smith, E.O., Rattner, Z., Gindi, G., Hoffer, P.B.: Computerized three-dimensional segmented human anatomy. Med. Phys. 21(2), 299–302 (1994)
Funding
Open Access funding provided by Budapest University of Technology and Economics
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This research has been financed by OTKA K–124124.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Szirmay-Kalos, L., Kacsó, Á., Magdics, M. et al. Robust compartmental model fitting in direct emission tomography reconstruction. Vis Comput 38, 655–668 (2022). https://doi.org/10.1007/s00371-020-02041-x
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00371-020-02041-x