Abstract
Background
Extracorporeal shock wave lithotripsy (ESWL) is considered one of the best choices for the treatment of various kinds of urinary tract calculi, although it might cause acute kidney injury.
Objective
To measure the urinary long non-coding RNA-messenger RNA (LncRNA-mRNA) panel before and after ESWL to evaluate post-ESWL renal injury in a reliable and non-invasive method.
Patients and methods
The study included 60 patients with renal stones treated with ESWL and 30 healthy volunteers. Voided urine samples were obtained before, 2 h, and 1 day after ESWL. We measured the urinary level of LncRNA (SBF2-AS1, FENDRR-19) and mRNA (GBP1, NLRP3) by real-time qPCR and compared the results with serum creatinine and eGFR.
Results
LncRNA (SBF2-AS1, FENDRR-19) and mRNA (GBP1, NLRP3) levels were higher in patients with renal stones when compared with healthy volunteers. They showed a statistically significant increase in the level of LncRNA-mRNA panel in baseline and after ESWL treatment.
Conclusion
LncRNA (SBF2-AS1, FENDRR-19) and mRNA (GBP1, NLRP3) levels were significantly elevated following ESWL treatment, highlighting the usefulness of urinary biomarkers in identifying patients at higher risk of developing renal injury after ESWL treatment.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The prevalence of urinary stones is around 3–12% and its recurrence rates can reach up to 50% within 10 years [1]. ESWL is considered the first line of treatment together with retrograde intrarenal surgery (RIRS) for stones smaller than 2 cm [2]. ESWL can cause the injury of thin-walled renal vessels, leading to transient haematoma, the release of cytokines/inflammatory cellular mediators and the infiltration of tissue by inflammatory response cells [3].
Serum creatinine and blood urea nitrogen levels, which are routinely used to detect and follow the progression of renal injury, are insensitive, nonspecific, and get higher only after significant kidney damage [4]. Till now, there is no consistent dependable urinary marker to permit the detection of acute kidney injury.
Inflammasome signaling regulates caspase-dependent inflammation and apoptosis. Several inflammasome-linked genes have a crucial role in kidney diseases, especially the NLRP3 (NOD-, LRR- and pyrin domain-containing 3) which contributes to many acute and chronic renal diseases [5]. Guanylate Binding Proteins (GBP) seem to play a critical role in inflammasome activation. They are expressed in immune cells and the stroma of the lung, kidney, and brain [6].
Circulating urinary long non-coding RNAs (LncRNAs) are fascinating novel biomarkers that reflect intra-nuclear processes noninvasively and may thus provide a better estimate of intracellular processes than currently established biomarkers [7]. LncRNAs are transcripts with a length of more than 200 nucleotides that exhibit tissue-specific expression and are involved in epigenetic regulation [8]. In several studies, circulating LncRNAs have been described as a fascinating new player in pathophysiological studies and the search for novel diagnostic and therapeutic strategies in kidney disease [9,10,11].
The four selected RNAs gene ontology is not linked only to inflammation, but also extended to different injury response patterns, e.g., renal apoptosis, GTPase activity, and ischemia reperfusion after ESWL induced kidney injury. Thus, their differential expression level not only reflect the inflammation in kidney injury, but also highlight their role in ischemic renal tissue and apoptotic tissues [12].
In this study, we hypothesized that an RNA panel linked to the inflammasome system and specific to kidney injury could be used as a potential biomarker panel, as combined LncRNA-mRNA panels are more informative than single RNA. We first identified inflammasome-related genes and their epigenetic regulators via in silico data analysis. Then, to confirm this panel, we assessed the differential expression of lncRNA [SBF2-AS1 (SET binding factor 2 antisense RNA1) and FENDRR-19 (Fetal-lethal non-coding developmental regulatory RNA)] and mRNA [NLRP3 (NOD-like receptor and pyrin domain-containing 3) and GBP1 (guanylate-binding protein 1)] in the urine of renal stone patients treated with ESWL, then we compared with healthy volunteers to evaluate their usefulness as diagnostic biomarkers for post ESWL kidney injury.
Patients and methods
The study was carried out after approval from Ain Shams Faculty’s Medicine Ethical Committee from December 2020 to July 2021. The participants were 60 patients whose gender and ages matched with 30 healthy volunteers with no history of kidney or stone disease. All patients underwent ESWL; for the first time to treat radiopaque stone(s) of 2 cm in diameter or less, located in the kidney. They were recruited from the outpatient clinic of the Urology Department of Ain Shams University hospitals. Informed consent was provided by all participants. The level of kidney disease was staged according to the National Kidney Foundation Kidney Disease Outcomes Quality Initiative Classification [13]. Patients with a history of active urinary tract infection, bleeding disorders, elevated serum creatinine, chronic renal failure (eGFR < 30) and pregnant females were excluded from the study (Supplementary Table 1S).
The procedure was done with 3500 shock waves and the frequency of shock waves was set at 60 shocks per minute.
Each patient provided three urine samples in centrifugal tubes (2 h before, then 2 h and 24 h after ESWL) and blood samples. Urine was then centrifuged at 4000 rpm for 10 min, and the urinary pellet was washed twice with phosphate-buffered saline. The resultant urine pellet was preserved at − 80 °C. Sera samples were collected and stored within 15 min at a temperature of − 80 °C.
We have selected the NLRP3 mRNA gene, which is highly correlates with inflammation and is essential for proper inflammasome formation and processing. Firstly, we selected an inflammatory pathway closely linked to kidney injury using biosystems available at the NCBI gene database (available at ncbi.nim.nih.gov/gene) (Supplementary Fig. 1S). Secondly, we selected the NLRP3 mRNA gene, which is closely linked to the inflammasome using the Reactome database (Supplementary Fig. 2S) (available at https://reactome.org/content/detail/R-HSA-844456).
Then, the gene’s ontology was verified (Supplementary Fig. 3S) (available at https://www.ncbi.nlm.nih.gov/gene?Db=gene&Cmd=DetailsSearch&Term=114548) followed by validating its basal expression in the kidney using the Gene Cards database (available at https://www.genecards.org/cgi-bin/carddisp.pl?gene=NLRP3) (Supplementary Fig. 4S). Thirdly, we selected SBF2-AS1 targeting NLRP3 mRNA using the “lncRNA2target” database (available at 123.59.132.21/lncrna2target/search.jsp) (Supplementary Fig. 5S). The selection of lncRNA is based on how strongly it interacts with mRNA, the novelty in kidney disease, and their basal expression in the kidney (available at https://www.genecards.org/cgi-bin/carddisp.pl?gene=SBF2-AS1&keywords=SBF2%5C-AS1) (Supplementary Fig. 6S).
Fourthly, we retrieved data on the GBP1 mRNA gene, which is involved in cytokine binding and inflammasome signaling, and was correlated with kidney injury (Supplementary Fig. 7S) (available at https://www.ncbi.nlm.nih.gov/gene/2633). We verified its expression in the Genecards database (available at genecards.org/cgi-bin/carddisp.pI?gene = GBP1&keywords = GBP1) (Supplementary Fig. 8S). Finally, we used the “lncRNA2target” database to find a related lncRNA LincFOXF1 (lncRNA-FENDRR:19) (ENSG00000268388) which is supposed to control the expression of the GBP1 gene (Supplementary Fig. 9S), followed by verifying its expression in the kidney (available at 123.59.132.21/lncrna2target/search.jsp) (Supplementary Fig. 10S).
Total RNA was extracted from the urine pellet using an miRNEasy RNA isolation kit (Qiagen, Hilden, Germany) according to the manufacturer's protocol. Total RNA samples were dissolved in 30 µl of nuclease-free water. Quality and quantity were checked using NanoDrop spectrophotometer. cDNA libraries for mRNAs, LncRNAs and miRNAs were prepared using the miScript II RT Kit (Qiagen, Germany). 4ul of 5 × miScript HiFlex Buffer, 2ul of 10 × miScript Nucleics Mix, 1ul of miScript Reverse Transcriptase Mix and RNase free water were added to 2ug of RNA extract, then incubated at 37 °C for 60 min and at 95 °C for 5 min using the Rotor-Gene thermal cycler (Thermo Electron Waltham, MA).
LncRNA (SBF2-AS1, FENDRR19) and mRNA (NLRP3, GBP1) expressions in urine samples of diseased groups and healthy control groups were quantified by qRT-PCR using QuantiTect SYBR-Green PCR Master Mix (Roche) and 10ul 2 × RT2 SYBR Green ROX qPCR Master mix. Sequentially, specific primers of each gene were designed.RT2 LncRNA qPCR Assay for Human LncRNA (SBF2-AS1, FENDRR19) and mRNA (NLRP3, GBP1) QuantiTect Primer Assay (NM_021202), RNase free water and 2ul template cDNA to a final volume of 20ul. Hs ACTB1SG QuantiTect Primer Assay (NM_001101) was used as a housekeeping gene in equalization of raw data like the invariant control for the samples and to compare with a reference sample. PCR programmed for relative LncRNA (SBF2-AS1, FENDRR19) quantification as follows: initial denaturation at 95 °C for 10 min; followed by 45 cycles at 95 °C for 15 s; then annealing at 55 °C for 30 s and extension at 70 °C for 30 s.
The real-time cycler was programmed as follows: initial activation step at 95 °C for 15 min to activate HotStarTaq DNA Polymerase. 40 cycles of PCR were performed under the following conditions: at 94 °C for 15 s, 55 °C for 30 s and 72 °C for 30 s for extension, denaturation and annealing sequentially. Each reaction was carried out three times. Fold change and expression levels were calculated using the 2 − ΔΔCt method. The Rotor-Gene real-time PCR detection system (Qiagen, Hilden, Germany) calculated the threshold cycle (Ct) value of each sample. The value was considered negative if higher than 36 Ct value. The amplification plot curve and melting curve were analyzed to confirm the specificities of the amplicons and Tm values.
The data were statistically presented using SPSS 20. Independent t-test, chi-square test, and Mann Whitney test were used. The (ROC) curve was done to characterize the predictive value of the selected RNA-based biomarker panel for post ESWEL kidney injury. The Spearman correlation was carried out to detect the association between clinicopathological parameters and RNA panel expression. A two-tailed p value of 0.05 or less was supposed to be statistically significant.
Results
In this study, there were no statistically significant differences between the two investigated groups regarding age, sex, body mass index, Serum Creatinine and eGFR and hypertension (p > 0.05).
Details of the demographic and clinical data are shown in (Table 1).
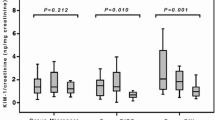
We reported that the positivity rates for urine LncRNA (SBF2-AS1, FENDRR19) and mRNA (GBP1, NLRP3) significantly increased in 24 h post-ESWL. Baseline pre-ESWL voided urine samples collected from patients with renal stones revealed significantly higher positive rates in comparison with the healthy volunteers’ voided urine (p < 0.01). We found that the four RNAs of all patients significantly increased in voided urinary specimens collected 2 and 24 h after ESWL compared with their pre-procedure baseline levels (Table 2, Supplementary Fig. 11S).
Urinary markers continued to rise significantly 1 day after ESWL (p < 0.01). Baseline pre-ESWL voided urine samples collected from patients with renal stones revealed significantly higher RNAs levels when compared to healthy volunteers’ voided urine (p < 0.01) (Supplementary Fig. 12S).
ROC curve analysis and the area under the curve (AUC) values were used to estimate the discriminative power of our selected RNAs between patients with renal stone versus the control group (as illustrated in Supplementary Fig. 13S).
Comparing the diseased groups with healthy control groups shows that the best discriminating cutoff values of LncRNA (SBF2-AS1, FENDRR19) and mRNA (GBP1, NLRP3) were 1.250, 1.250, 1.250 and 1.360, respectively. The sensitivities measured 91.7%, 76.7%, 78.3% and 78.3%, respectively. Accordingly, this result indicates that these thresholds could be used to differentiate/identify diseased patients from healthy subjects (Supplementary Table 2S).
There was a highly significant correlation between all the investigated urine RNAs and serum creatinine. Also, there was a highly significant correlation between serum eGFR and FENDRR19, as well as a significant correlation between serum eGFR and SBF2-AS1.
By studying the correlation between the studied RNA-based biomarkers, we found that there was a highly significant positive correlation between all of them [(LncRNA (SBF2-AS1, FENDRR19) and mRNA (GBP1, NLRP3)] based on fold changes (R.Q.) among all the study groups. Results are shown in Supplementary Table 3S.
No baseline characteristic variables were found to have a significant association by the multilinear regression model. All the tested predictor variables are presented in Supplementary Table 4S. mRNA-NLRP3 and serum creatinine is the most significant predictor of kidney injury (p = 0.000,0.002, respectively).
Discussion
ESWL subjects the renal parenchyma to high levels of energy, leading to broad spectrum of vascular kidney damage ranging from self-limited hematuria to perinephric/nephric hematomas [11]. In the long term, 11 animal and 13 human studies have suggested that these acute hemorrhagic lesions may progress to scar formation and complete the atrophy of the renal papillae. There is no existing adequate imaging modality available to assess the parenchymal injury, thus creating a need for a potential novel diagnostic test that can reliably detect such renal injuries [14].
We identified that the level of expression of LncRNA (SBF2-AS1, FENDRR19) and mRNA (GBP1, NLRP3)) are highly detected in the urine of post ESWL-procedure patients. This has raised the possibility of using this network as a circulating biomarker for post ESWL renal injury detection. Also, as a part of inflammation, the results of this study revealed that urine NLRP3-mRNA was significantly upregulated in diseased groups compared to normal healthy individuals in the controlled groups (p < 0.01), and it is differentially expressed after overexpression of urine lncRNA SBF2-AS1. Previous reports on cell lines and tissue also indicated that interfering with the process of NLRP3 inflammasome activation can regulate kidney injury [15]. Furthermore, activation of NLRP3 inflammatory corpuscles could promote AKI induced by sepsis. Simultaneously, a renal injury may lead to the production of mitochondrial reactive oxygen species (mROS), which may induce the binding of TXNIP to NLRP3 Moreo [16].
With regards to GBP1-mRNA, the results of our study revealed that its expression was significantly upregulated in AKI patients compared to healthy individuals (p < 0.01) and it is differentially expressed after overexpression of urine lncRNA FENDRR19. Honkala et al. declared that GBP1 governs cellular responses to infection, inflammation, and environmental stressors [17]. At the cellular level, GBP1 activation both restrains proliferation and protects against apoptosis in inflammatory contexts [18].
LncRNAs play a critical role in immunity as they regulate the survival, differentiation and cytokine formation of immune cells [19]. LncRNAs may participate in epigenetic regulation for proinflammatory and anti-inflammatory gene expression in macrophages challenged by inflammatory mediators [20]. Overexpression of lncRNA genes or deficiency has been involved in kidney diseases; they not only function as biomarkers, but also as pathogenic mediators of kidney diseases. Thus, LncRNAs associated with kidney disease identification and characterization may provide new diagnostic and therapeutic opportunities for renal disorders [21].
FENDRR lncRNA (Foxf1 adjacent non-coding developmental regulatory RNA) plays a significant role in heart development [22]. Çekin and his colleagues found that FENDRR expression was lower in coronary artery disease [23]. Interestingly, Munteanu et al.’s results suggested that FENDRR promoted polarization of M1 macrophage and so, targeting FENDRR may act as a potential therapeutic target for the treatment of diseases that occurred with polarization macrophage [24].
Of note, lncRNA SBF2-AS1 is located at the chromosome 11p15.1 locus. Several studies showed that lncRNA SBF2-AS1 might act as an essential regulator of tumor progression [25,26,27]. LncRNA SBF2-AS1 induces hepatocellular carcinoma metastasis by regulating epithelial-mesenchymal transition [28].
The results of our study revealed that urinary LncRNA (SBF2-AS1, FENDRR19) and mRNA (GBP1, NLRP3) were significantly upregulated in AKI patients when compared to people in healthy control groups. Interestingly, pre ESWL level of urinary SBF2-AS1; and to lesser extent the rest of urinary RNAs measured, is significantly higher than that of healthy control. This can be attributed to kidney injury because of stone obstruction. Followed by sharp rise of the 4 RNA markers 24 h post ESWL compared to pre ESWL level in the patients group.
Accuracy of post ESWL renal injury detection can be improved by measuring urine LncRNA (SBF2-AS1, FENDRR19) and mRNA (GBP1, NLRP3). Pointedly, there was a highly significant positive correlation between urine [LncRNA (SBF2-AS1, FENDRR19) and mRNA (GBP1, NLRP3)] based on fold changes (R.Q.s) among the human study groups.
We have tried to identify baseline characteristic variables that could identify risky patients with significant renal injury after ESWL. We performed a univariate and multivariate analysis. Serum creatinine, Lnc-RNA-FENDRR-19 and NLRP3 mRNA were significant independent variables that significantly correlated with renal injury.
Limitations of the study include its inclusion of a relatively small sample size. Moreover, in vitro and in vivo functional analyses are needed to clarify the biological mechanisms of RNA-RNA crosstalk in post ESWL renal injury by assessment of the selected RNAs in rat AKI animal model for further verification.
Conclusion
Urine LncRNA (SBF2-AS1, FENDRR19) and mRNA (GBP1, NLRP3) rise significantly post ESWL. Hence, they may be prospective clinical markers for assessing acute renal injury following ESWL.
References
Preminger GM, Tiselius HG, Assimos DG, Alken P, Buck C, Gallucci M, Knoll T, Lingeman JE, Nakada SY, Pearle MS, Sarica K (2007) guideline for the management of ureteral calculi. J Urol 178(6):2418–2434
Türk C, Petřík A, Sarica K, Seitz C, Skolarikos A, Straub M, Knoll T (2016) EAU guidelines on interventional treatment for urolithiasis. Eur Urol 69(3):475–482
Connors BA, Evan AP, Blomgren PM, Hsi RS, Harper JD, Sorensen MD, Wang YN, Simon JC, Paun M, Starr F, Cunitz BW (2014) Comparison of tissue injury from focused ultrasonic propulsion of kidney stones versus extracorporeal shock wave lithotripsy. J Urol 191(1):235–241
Bryniarski P, Paradysz A, Zyczkowski M, Kupilas A, Nowakowski K, Bogacki R (2012) A randomized controlled study to analyze the safety and efficacy of percutaneous nephrolithotripsy and retrograde intrarenal surgery in the management of renal stones more than 2 cm in diameter. J Endourol 26(1):52–57
Komada T, Muruve DA (2019) The role of inflammasomes in kidney disease. Nat Rev Nephrol 15(8):501–520
Kim BH, Chee JD, Bradfield CJ, Park ES, Kumar P, MacMicking JD (2016) Interferon-induced guanylate-binding proteins in inflammasome activation and host defense. Nat Immunol 17(5):481–489
Martens-Uzunova ES, Böttcher R, Croce CM, Jenster G, Visakorpi T, Calin GA (2014) Long noncoding RNA in prostate, bladder, and kidney cancer. Eur Urol 65(6):1140–1151
Cabili MN, Trapnell C, Goff L, Koziol M, Tazon-Vega B, Regev A, Rinn JL (2011) Integrative annotation of human large intergenic noncoding RNAs reveals global properties and specific subclasses. Genes Dev 25(18):1915–1927
Wang X, Xu Y, Zhu YC, Wang YK, Li J, Li XY, Ji T, Bai SJ (2019) LncRNA NEAT1 promotes extracellular matrix accumulation and epithelial-to-mesenchymal transition by targeting miR-27b-3p and ZEB1 in diabetic nephropathy. J Cell Physiol 234(8):12926–12933
Li Y, Ren D, Xu G (2019) Long noncoding RNA MALAT1 mediates high glucose-induced glomerular endothelial cell injury by epigenetically inhibiting klotho via methyltransferase G9a. IUBMB Life 71(7):873–881
Ignarski M, Islam R, Müller RU (2019) Long non-coding RNAs in kidney disease. Int J Mol Sci 20(13):3276
Wu YL, Li HF, Chen HH, Lin H (2020) MicroRNAs as biomarkers and therapeutic targets in inflammation-and ischemia-reperfusion-related acute renal injury. Int J Mol Sci 21(18):6738
Levey AS, Coresh J, Bolton K, Culleton B, Harvey KS, Ikizler TA, Johnson CA, Kausz A, Kimmel PL, Kusek J, Levin A (2002) K/DOQI clinical practice guidelines for chronic kidney disease: evaluation, classification, and stratification. Am J Kidney Dis 39(2 Suppl 1):i-ii+
Fahmy N, Sener A, Sabbisetti V, Nott L, Lang RM, Welk BK, Méndez-Probst CE, MacPhee RA, VanEerdewijk S, Cadieux PA, Bonventre JV (2013) Urinary expression of novel tissue markers of kidney injury after ureteroscopy, shockwave lithotripsy, and in normal healthy controls. J Endourol 27(12):1455–1462
Xiang H, Zhu F, Xu Z, Xiong J (2020) Role of inflammasomes in kidney diseases via both canonical and non-canonical pathways. Frontiers in cell and developmental biology 8:106
Deng H, Chen F, Wang Y, Jiang H, Dong Z, Yuan B, Zhao X (2020) The role of activated NLRP3 inflammatory body in acute kidney injury in rats caused by sepsis and NLRP3-TXNIP signaling pathway. Saudi J Biol Sci 27(5):1251–1259
Honkala AT, Tailor D, Malhotra SV (2020) Guanylate-binding protein 1: an emerging target in inflammation and cancer. Front Immunol 10:3139
Naschberger E, Bauer M, Stürzl M (2006) Human guanylate binding protein-1 (hGBP-1) characterizes and establishes a non-angiogenic. Adv Enzyme Regul 45:215
Chen YG, Satpathy AT, Chang HY (2017) Gene regulation in the immune system by long noncoding RNAs. Nat Immunol 18(9):962–972
Liu J, Lao L, Chen J, Li J, Zeng W, Zhu X, Li J, Chen X, Yang L, Xing Y, Chen F (2021) The IRENA lncRNA converts chemotherapy-polarized tumor-suppressing macrophages to tumor-promoting phenotypes in breast cancer. Nat Cancer 2(4):457–473
Zhou Q, Chen W, Yu XQ (2020) Long non-coding RNAs as novel diagnostic and therapeutic targets in kidney disease. Chronic Dis Transl Med 5(4):252–257. https://doi.org/10.1016/j.cdtm.2019.12.004
Grote P, Herrmann BG (2013) The long non-coding RNA Fendrr link epigenetic control mechanisms to gene regulatory networks in mammalian embryogenesis. RNA Biol 10(10):1579–1585
Çekin N, Özcan A, Göksel S, Arslan S, Pınarbaşı E, Berkan Ö (2018) Decreased FENDRR and LincRNA-p21 expression in atherosclerotic plaque. Anatol J Cardiol 19(2):131
Munteanu MC, Huang C, Liang Y, Sathiaseelan R, Zeng X, Liu L (2020) Long non-coding RNA FENDRR regulates IFNγ-induced M1 phenotype in macrophages. Sci Rep 10(1):1–12
Gao F, Feng J, Yao H, Li Y, Xi J, Yang J (2019) LncRNA SBF2-AS1 promotes the progression of cervical cancer by regulating the miR-361-5p/FOXM1 axis. Artif Cells Nanomed Biotechnol 47(1):776–782
Zhao QS, Li L, Zhang L, Meng XW, Li LL, Ge XF, Li ZP (2016) Over-expression of lncRNA SBF2-AS1 is associated with advanced tumor progression and poor prognosis in patients with non-small cell lung cancer. Eur Rev Med Pharmacol Sci 20(14):3031–3034
Lv J, Qiu M, Xia W, Liu C, Xu Y, Wang J, Leng X, Huang S, Zhu R, Zhao M, Ji F (2016) High expression of long non-coding RNA SBF2-AS1 promotes proliferation in non-small cell lung cancer. J Exp Clin Cancer Res 35(1):1–13
Shi ZG, Sun Y, Wang KS, Jia JD, Yang J, Li YN (2019) Effects of miR-26a/miR-146a/miR-31 on airway inflammation of asthma mice and asthma children. Eur Rev Med Pharmacol Sci 23(12):5432–5440
Funding
Open access funding provided by The Science, Technology & Innovation Funding Authority (STDF) in cooperation with The Egyptian Knowledge Bank (EKB). This work was supported by Science, Technology and Innovation Funding Agency (STDF), Project No. 28889.
Author information
Authors and Affiliations
Contributions
AT: Conceptualization, data collection, fund acquisition, methodology. MM: Conceptualization, methodology, data curation, formal analysis, funding acquisition, data analysis, manuscript editing. SS: Data analysis, Statics, manuscript writing, investigation. SHAA: Data curation, investigation. MS: Data analysis, investigation. HS: Conceptualization, funding acquisition, methodology. MMYS: Investigation, ESWL procedure. MSS: Funding acquisition, investigation, ESWL procedure. AR: Investigation, ESWL procedure. AAS: Investigation, ESWL procedure. WM: Conceptualization, project development, fund acquisition, manuscript editing, corresponding author.
Corresponding authors
Ethics declarations
Conflict of interest
All the authors declare no competing interests.
Ethical approval
Written informed consent was obtained from all the participants of this study, which was performed following the Declaration of Helsinki, and was approved by the Research Ethics Committee of Ain Shams University, Faculty of Medicine, Egypt.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Tawfick, A., Matboli, M., Shamloul, S. et al. Predictive urinary RNA biomarkers of kidney injury after extracorporeal shock wave lithotripsy. World J Urol 40, 1561–1567 (2022). https://doi.org/10.1007/s00345-022-03996-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00345-022-03996-3