Abstract
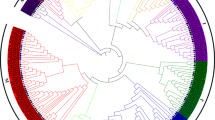
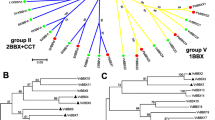
Basic leucine zipper (bZIP) transcription factor family members play important roles in many aspects of anthocyanin accumulation and adaptation to the environment. However, little information is available about the bZIP genes in black raspberry. Herein, 49 RobZIP genes were identified and renamed based on their respective chromosome distribution (including five that could not be located to chromosomes). According to phylogenetic analysis, RobZIPs are divided into 13 subfamilies. RobZIPs have 1–19 exons, 137–890 amino acids, and theoretical PIs of 4.79–10.3. Moreover, most RobZIP genes are located in the nucleus. The analysis of cis-acting elements in the promoters of these genes showed that there were many elements related to light, hormones and abiotic stress. Through abscisic acid (ABA) treatment, we found that most RobZIP genes in black raspberry fruits or leaves were significantly expressed, but the trends in the responses to ABA differed. In addition, there were differences in the expression levels of RobZIP genes in different tissues and fruit developmental stages of black raspberry, which suggest possible different functions of RobZIP members. This study identified and analyzed the bZIP family genes in black raspberry to provide a basis for further functional verification of bZIP genes.






Similar content being viewed by others
References
An JP, Qu FJ, Yao JF, Wang XN, You CX, Wang XF, Hao YJ (2017) The bZIP transcription factor MdHY5 regulates anthocyanin accumulation and nitrate assimilation in apple. Hortic Res 4:17023. https://doi.org/10.1038/hortres.2017.56
An JP, Yao JF, Xu RR, You CX, Wang XF, Hao YJ (2018) Apple bZIP transcription factor MdbZIP44 regulates abscisic acid-promoted anthocyanin accumulation. Plant Cell Environ 41:2678–2692. https://doi.org/10.1111/pce.13393
Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS (2009) MEME SUITE: Tools for motif discovery and searching. Nucleic Acids Res 37:W202–W208. https://doi.org/10.1093/nar/gkp335
Banerjee A, Roychoudhury A (2017) Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma 254:3–16. https://doi.org/10.1007/s00709-015-0920-4
Bi C, Yu Y, Dong C, Yang Y, Zhai Y, Du F, Xia C, Ni Z, Kong X, Zhang L (2021) The bZIP transcription factor TabZIP15 improves salt stress tolerance in wheat. Plant Biotechnol J 19:209–211. https://doi.org/10.1111/pbi.13453
Chen S, Ma T, Song S, Li X, Fu P, Wu W, Liu J, Gao Y, Ye W, Dry IB et al (2021) Arabidopsis downy mildew effector HaRxLL470 suppresses plant immunity by attenuating the DNA-binding activity of bZIP transcription factor HY5. New Phytol 230:1562–1577. https://doi.org/10.1111/nph.17280
Corrêa LGG, Riaño-Pachón DM, Schrago CG, dos Santos RV, Mueller-Roeber B, Vincentz M (2008) The role of bZIP transcription factors in green plant evolution: Adaptive features emerging from four founder genes. PLoS One 3:e2944. https://doi.org/10.1371/journal.pone.0002944
Dröge-Laser W, Snoek BL, Snel B, Weiste C (2018) The Arabidopsis bZIP transcription factor family — an update. Curr Opin Plant Biol 45:36–49. https://doi.org/10.1016/j.pbi.2018.05.001
Ellenberger TE, Brandl CJ, Struhl K, Harrison SC (1992) The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α Helices: Crystal structure of the protein-DNA complex. Cell 71:1223–1237. https://doi.org/10.1016/s0092-8674(05)80070-4
Feng Y, Wang Y, Zhang G, Gan Z, Gao M, Lv J, Wu T, Zhang X, Xu X, Yang S et al (2021) Group-C/S1 bZIP hetero-dimers regulate MdIPT5b to negatively modulate drought tolerance in apple species. Plant J 107:399–417. https://doi.org/10.1111/tpj.15296
Foster TM, Bassil NV, Dossett M, Leigh Worthington M, Graham J (2019) Genetic and genomic resources for Rubus breeding: A roadmap for the future. Hortic Res 6:116. https://doi.org/10.1038/s41438-019-0199-2
Furihata T, Maruyama K, Fujita Y, Umezawa T, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2006) Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc Natl Acad Sci USA 103:1988–1993. https://doi.org/10.1073/pnas.0505667103
Hirayama T, Shinozaki K (2007) Perception and transduction of abscisic acid signals: Keys to the function of the versatile plant hormone ABA. Trends Plant Sci 12:343–351. https://doi.org/10.1016/j.tplants.2007.06.013
Hu W, Yang H, Yan Y, Wei Y, Tie W, Ding Z, Zuo J, Peng M, Li K (2016) Genome-wide characterization and analysis of bZIP transcription factor gene family related to abiotic stress in cassava. Sci Rep 6:22783. https://doi.org/10.1038/srep22783
Jain M, Khurana P, Tyagi AK, Khurana JP (2008) Genome-wide analysis of intronless genes in rice and Arabidopsis. Funct Integr Genomics 8:69–78. https://doi.org/10.1007/s10142-007-0052-9
Jakoby M, Weisshaar B, Dröge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, Parcy F (2002) bZIP transcription factors in Arabidopsis. Trends Plant Sci 7:106–111. https://doi.org/10.1016/s1360-1385(01)02223-3
Jibran R, Dzierzon H, Bassil N, Bushakra JM, Edger PP, Sullivan S, Finn CE, Dossett M, Vining KJ, VanBuren R et al (2018) Chromosome-scale scaffolding of the black raspberry (Rubus occidentalis L.) genome based on chromatin interaction data. Hortic Res. https://doi.org/10.1038/s41438-017-0013-y
Landschulz WH, Johnson PF, McKnight SL (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240:1759–1764. https://doi.org/10.1126/science.3289117
Latchman DS (1997) Transcription factors: an overview. Int J Biochem Cell Biol 29:1305–1312. https://doi.org/10.1016/s1357-2725(97)00085-x
Lee SC, Choi HW, Hwang IS, Choi DS, Hwang BK (2006) Functional roles of the pepper pathogen-induced bZIP transcription factor, CAbZIP1, in enhanced resistance to pathogen infection and environmental stresses. Planta 224:1209–1225. https://doi.org/10.1007/s00425-006-0302-4
Li D, Fu F, Zhang H, Song F (2015) Genome-wide systematic characterization of the bZIP transcriptional factor family in tomato (Solanum lycopersicum L.). BMC Genom 16:60–78. https://doi.org/10.1186/s12864-015-1990-6
Li YY, Meng D, Li M, Cheng L (2016) Genome-wide identification and expression analysis of the bZIP gene family in apple (Malus domestica). Tree Genet Genomes 12:1–17. https://doi.org/10.1007/s11295-016-1043-6
Liang Y, Xia J, Jiang Y, Bao Y, Chen H, Wang D, Zhang D, Yu J, Cang J (2022) Genome-wide identification and analysis of bZIP gene family and resistance of TaABI5 (TabZIP96) under freezing stress in wheat (Triticum aestivum). Int J Mol Sci 23:2351. https://doi.org/10.3390/ijms23042351
Liu J, Chen N, Chen F, Cai B, Dal Santo S, Tornielli GB, Pezzotti M, Cheng ZMM (2014) Genome-wide analysis and expression profile of the bZIP transcription factor gene family in grapevine (Vitis vinifera). BMC Genom 15:1–18. https://doi.org/10.1186/1471-2164-15-281
Nijhawan A, Jain M, Tyagi AK, Khurana JP (2008) Genomic survey and gene expression analysis of the basic leucine zipper transcription factor family in rice. Plant Physiol 146:333–350. https://doi.org/10.1104/pp.107.112821
Nuruzzaman M, Manimekalai R, Sharoni AM, Satoh K, Kondoh H, Ooka H, Kikuchi S (2010) Genome-wide analysis of NAC transcription factor family in rice. Gene 465:30–44. https://doi.org/10.1016/j.gene.2010.06.008
Oliva R, Ji C, Atienza-Grande G, Huguet-Tapia JC, Perez-Quintero A, Li T, Eom JS, Li C, Nguyen H, Liu B, Auguy F, Sciallano C, Luu VT, Dossa GS, Cunnac S, Schmidt SM, Slamet-Loedin IH, Vera Cruz C, Szurek B, Frommer WB, White FF, Yang B (2019) Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat Biotechnol 37:1344–1350. https://doi.org/10.1038/s41587-019-0267-z
Pantelidis G, Vasilakakis M, Manganaris G, Diamantidis G (2007) Antioxidant capacity, phenol, anthocyanin and ascorbic acid contents in raspberries, blackberries, red currants, gooseberries and Cornelian cherries. Food Chem 102:777–783. https://doi.org/10.1016/j.foodchem.2006.06.021
Rogozin IB, Sverdlov AV, Babenko VN, Koonin EV (2005) Analysis of evolution of exon-intron structure of eukaryotic genes. Brief Bioinform 6:118–134. https://doi.org/10.1093/bib/6.2.118
Schlögl PS, Fábio TS, Nogueira RD, Felix JM, De Rosa VE, Vicentini R, Leite A, Ulian EC, Menossi M (2008) Identification of new ABA- and MEJA-activated sugarcane bZIP genes by data mining in the SUCEST database. Plant Cell Rep. 27:335–345. https://doi.org/10.1007/s00299-007-0468-7
Sirichandra C, Davanture M, Turk BE, Zivy M, Valot B, Leung J, Merlot S (2010) The Arabidopsis ABA-activated kinase OST1 phosphorylates the bZIP transcription factor ABF3 and creates a 14–3–3 binding site involved in its turnover. PLoS One 5:e13935. https://doi.org/10.1371/journal.pone.0013935
Sornaraj P, Luang S, Lopato S, Hrmova M (2016) Basic leucine zipper (bZIP) transcription factors involved in abiotic stresses: a molecular model of a wheat bZIP factor and implications of its structure in function. Biochim. Biophys. Acta (BBA) - Gen. Subj 1860:46–56. https://doi.org/10.1016/j.bbagen.2015.10.014
Talanian RV, McKnight CJ, Kim PS (1990) Sequence-specific DNA binding by a short peptide dimer. Science 249:769–771. https://doi.org/10.1126/science.2389142
Tao YT, Chen LX, Jin J, Du ZK, Li JM (2022) Genome-wide identification and analysis of bZIP gene family reveal their roles during development and drought stress in Wheel Wingnut (Cyclocarya paliurus). BMC Genom 23:743. https://doi.org/10.1186/s12864-022-08978-8
Uno Y, Furihata T, Abe H, Yoshida R, Shinozaki K, Yamaguchi-Shinozaki K (2000) Arabidopsis basic leucine zipper transcription factors involved in an abscisic acid-dependent signal transduction pathway under drought and high-salinity conditions. Proc Natl Acad Sci U S A 97:11632–11637. https://doi.org/10.1073/pnas.190309197
van Buren R, Bryant D, Bushakra JM, Vining KJ, Edger PP, Rowley ER, Priest HD, Michael TP, Lyons E, Fil-ichkin SA et al (2016) The genome of black raspberry (Rubus occidentalis). Plant J 87:535–547. https://doi.org/10.1111/tpj.13215
Wang SY, Lin HS (2000) Antioxidant activity in fruits and leaves of blackberry, raspberry, and strawberry varies with cultivar and developmental stage. J Agric Food Chem 48:140–146. https://doi.org/10.1021/jf9908345
Wang XL, Chen X, Yang TB, Cheng Q, Cheng ZM (2017) Genome-wide identification of bZIP family genes involved in drought and heat stresses in strawberry (Fragaria vesca). Int J Genomics 2017:3981031. https://doi.org/10.1155/2017/3981031
Wang Z, Yan L, Wan L, Huai D, Kang Y, Shi L, Jiang H, Lei Y, Liao B (2019) Genome-wide systematic characterization of bZIP transcription factors and their expression profiles during seed development and in response to salt stress in peanut. BMC Genom 20:51. https://doi.org/10.1186/s12864-019-5434-6
Wang Z, Zhu J, Yuan W, Wang Y, Hu P, Jiao C, Xia H, Wang D, Cai Q, Li J et al (2021) Genome-wide characterization of bZIP transcription factors and their expression patterns in response to drought and salinity stress in Jatropha curcas. Int J Biol Macromol 181:1207–1223. https://doi.org/10.1016/j.ijbiomac.2021.05.027
Wei K, Chen J, Wang Y, Chen Y, Chen S, Lin Y, Pan S, Zhong X, Xie D (2012) Genome-wide analysis of bZIP-encoding genes in maize. DNA Res 19:463–476. https://doi.org/10.1093/dnares/dss026
Wu Y, Zhang C, Wu W, Li W, Lyu L (2021) Genome-wide identification and analysis of the MADS-box gene family and its potential role in fruit ripening in black raspberry (Rubus occidentalis L.). J Berry Res 11:301–315. https://doi.org/10.3233/jbr-200679
Wu Y, Zhang C, Huang Z, Lyu L, Li J, Li W, Wu W (2021) The color difference of rubus fruits is closely related to the composition of flavonoids including anthocyanins. LWT-Food Sci Technol. https://doi.org/10.1016/j.lwt.2021.111825
Wu Y, Zhang S, Huang X, Lyu L, Li W, Wu W (2022) Genome-wide identification of WRKY gene family members in black raspberry and their response to abiotic stresses. Sci Hortic 304:111338. https://doi.org/10.1016/j.scienta.2022.111338
Xu Z, Ali Z, Xu L, He X, Huang Y, Yi J, Shao H, Ma H, Zhang D (2016) The nuclear protein GmbZIP110 has transcription activation activity and plays important roles in the response to salinity stress in soybean. Sci Rep 6:2045–2057. https://doi.org/10.1038/srep20366
Yoshida T, Fujita Y, Sayama H, Kidokoro S, Maruyama K, Mizoi J, Shinozaki K, Yamaguchi-Shinozaki K (2010) AREB1, AREB2, and ABF3 are master transcription factors that cooperatively regulate ABRE-dependent ABA signaling involved in drought stress tolerance and require ABA for full activation. Plant J 61:672–685. https://doi.org/10.1111/j.1365-313X.2009.04092.x
Zhang Y, Zhang G, Xia N, Wang XJ, Huang LL, Kang ZS (2009) Cloning and characterization of a bZIP transcription factor gene in wheat and its expression in response to stripe rust pathogen infection and abiotic stresses. Physiol Mol Plant Pathol 73:88–94. https://doi.org/10.1016/j.pmpp.2009.02.002
Zhang H, Zhu J, Gong Z, Zhu J (2021) Abiotic stress responses in plants. Nat Rev Genet 23:104–119. https://doi.org/10.1038/s41576-021-00413-0
Zhao K, Chen S, Yao W, Cheng Z, Zhou B, Jiang T (2021) Genome-wide analysis and expression profile of the bZIP gene family in poplar. BMC Plant Biol 21:122. https://doi.org/10.1186/s12870-021-02879-w
Zhu JK (2016) Abiotic stress signaling and responses in plants. Cell 167:313–324. https://doi.org/10.1016/j.cell.2016.08.029
Zou M, Guan Y, Ren H, Zhang F, Chen F (2008) A bZIP transcription factor, OsABI5, is involved in rice fertility and stress tolerance. Plant Mol Biol 66:675–683. https://doi.org/10.1007/s11103-008-9298-4
Funding
This research was supported by the “JBGS” Project of Seed Industry Revitalization in Jiangsu Province (JBGS[2021]021), the earmarked fund for Jiangsu Agricultural Industry Technology System (JATS[2022]510), the Central Finance Forestry Technology Promotion and Demonstration Project (SU[2021]TG08), and the Jiangsu Institute of Botany Talent Fund (JIBTF202105).
Author information
Authors and Affiliations
Contributions
Yaqiong Wu designed the study and performed the experiments; Yaqiong Wu, Xin Huang and Chunhong Zhang performed the experiments; Yaqiong Wu and Lianfei Lyu analyzed the data; Yaqiong Wu wrote the manuscript; and Weilin Li and Wenlong Wu revised the manuscript. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of Interest
The authors declare no conflict of interest.
Additional information
Handling Editor: Raul Herrera.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Wu, Y., Huang, X., Zhang, C. et al. Genome-Wide Characterization and Expression of the bZIP Family in Black Raspberry. J Plant Growth Regul 43, 259–271 (2024). https://doi.org/10.1007/s00344-023-11082-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-023-11082-0