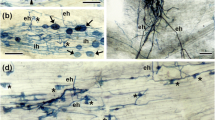
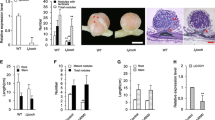
Abstract
Nodulation in legumes is regulated to maintain carbon–nitrogen homeostasis. A systemic negative feedback pathway, Autoregulation of Nodulation (AON) controls nodule number by suppressing further nodulation as early as 48–72 h post inoculation. In order to identify new genes involved in AON, a screen for suppressors of sunn-1, a Medicago truncatula AON mutant with a hypernodulation phenotype, was performed and identified a gene not previously associated with signaling in the legume-rhizobia symbiosis. We discovered that sunn-1 plants carrying the mutation suppressor of sunn-1 (sos1) displayed wild-type nodule numbers. At 3 days post inoculation (dpi), sos1 sunn-1 plants make many nodulation foci and infection threads but lack the emerging nodules observed in wild-type and sunn-1 mutants. At 10 dpi, in contrast to sunn-1, no significance in nodule number was observed between wildtype and the sos1 sunn-1plants. Using grafting, we showed that sos1 suppression of the sunn-1 nodule phenotype is dependent on the genotype of the root. The lesion a C to T transition resulting in an R to H a change mapped to Mediator 16A (MED16A) and expression of this gene in composite hairy roots led to recovery of hypernodulation in sos1 sunn-1plants. Induction of expression of the MtNIN transcription factor involved in regulating nodulation is reduced in sos1 sunn-1compared to sunn-1. Furthermore, sos1 plants display fewer nodules than wild-type but higher arbuscular density when colonized by an arbuscular mycorrhizal fungus, and shorter roots in the absence of rhizobia. Collectively, our findings show that effects of mutation of Mediator 16A are not specific for symbiosis but involve multiple root response pathways, making the sos1 mutant an important tool for dissection of regulons controlling rhizobial and mycorrhizal responses, root growth and possibly other response pathways.










Similar content being viewed by others
References
Abe A, Kosugi S, Yoshida K, Natsume S, Takagi H, Kanzaki H, Matsumura H, Yoshida K, Mitsuoka C, Tamiru M (2012) Genome sequencing reveals agronomically important loci in rice using MutMap. Nat Biotechnol 30:174. https://doi.org/10.1038/nbt.2095
Amor BB, Shaw SL, Oldroyd GE, Maillet F, Penmetsa RV, Cook D, Long SR, Dénarié J, Gough C (2003) The NFP locus of Medicago truncatula controls an early step of Nod factor signal transduction upstream of a rapid calcium flux and root hair deformation. Plant J 34:495–506. https://doi.org/10.1046/j.1365-313X.2003.01743.x
Andrews S (2010) FastQC. A quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/.
Boivin C, Camut S, Malpica CA, Truchet G, Rosenberg C (1990) Rhizobium meliloti genes encoding catabolism of trigonelline are induced under symbiotic conditions. Plant Cell 2:1157–1170. https://doi.org/10.1105/tpc.2.12.1157
Charpentier M, Sun J, Vaz Martins T, Radhakrishnan GV, Findlay K, Soumpourou E, Thouin J, Very AA, Sanders D, Morris RJ, Oldroyd GE (2016) Nuclear-localized cyclic nucleotide-gated channels mediate symbiotic calcium oscillations. Science 352:1102–1105. https://doi.org/10.1126/science.aae0109
Clèries R, Galvez J, Espino M, Ribes J, Nunes V, de Heredia ML (2012) BootstRatio: a web-based statistical analysis of fold-change in qPCR and RT-qPCR data using resampling methods. Comput Biol Med 42:438–445. https://doi.org/10.1016/j.compbiomed.2011.12.012
Crawford T, Karamat F, Lehotai N, Rentoft M, Blomberg J, Strand A, Björklund S (2020) Specific functions for mediator complex subunits from different modules in the transcriptional response of Arabidopsis thaliana to abiotic stress. Scientific Rep 10:5073. https://doi.org/10.1038/s41598-020-61758-wedess
Crook AD, Schnabel EL, Frugoli JA (2016) The systemic nodule number regulation kinase SUNN in Medicago truncatula interacts with MtCLV2 and MtCRN. Plant J 88:108–119
Dolan WL, Chapple C (2017) Conservation and divergence of mediator structure and function: insights from plants. Plant Cell Physiol 58:04–21. https://doi.org/10.1093/pcp/pcw176
Ferguson BJ, Indrasumunar A, Hayashi S, Lin M, Lin Y, Reid DE, Gresshoff PM (2010) Molecular analysis of legume nodule development and autoregulation. J Integr Plant Biol 52:61–76. https://doi.org/10.1111/j.1744-7909.2010.00899.x
Gonzalez-Rizzo S, Crespi M, Frugier F (2006) The Medicago truncatula CRE1 cytokinin receptor regulates lateral root development and early symbiotic interaction with Sinorhizobium meliloti. Plant Cell 18:2680–2693. https://doi.org/10.1105/tpc.106.043778
Guillotin B, Couzigou J, Combier J (2016) NIN is involved in the regulation of arbuscular mycorrhizal symbiosis. Front Plant Sci 7:1704. https://doi.org/10.3389/fpls.2016.01704
Hemsley PA, Hurst CH, Kaliyadasa E, Lamb R, Knight MR, De Cothi EA, Steele JF, Knight H (2014) The Arabidopsis mediator complex subunits MED16, MED14, and MED2 regulate mediator and RNA polymerase II recruitment to CBF-responsive cold-regulated genes. Plant Cell 26:465–484. https://doi.org/10.1105/tpc.113.117796
Hirsch S, Kim J, Munoz A, Heckmann AB, Downie JA, Oldroyd GE (2009) GRAS proteins form a DNA binding complex to induce gene expression during nodulation signaling in Medicago truncatula. Plant Cell 21:545–557. https://doi.org/10.1105/tpc.108.064501
Imin N, Mohd-Radzman NA, Ogilvie HA, Djordjevic MA (2013) The peptide-encoding CEP1 gene modulates lateral root and nodule numbers in Medicago truncatula. J Exp Bot 64:5395–5409. https://doi.org/10.1093/jxb/ert369
Jin Y, Liu H, Luo D, Yu N, Dong W, Wang C, Zhang X, Dai H, Yang J, Wang E (2016) DELLA proteins are common components of symbiotic rhizobial and mycorrhizal signalling pathways. Nat Commun 7:1–14. https://doi.org/10.1038/ncomms12433
Kakar K, Wandrey M, Czechowski T, Gaertner T, Scheible W, Stitt M, Torres-Jerez I, Xiao Y, Redman JC, Wu HC (2008) A community resource for high-throughput quantitative RT-PCR analysis of transcription factor gene expression in Medicago truncatula. Plant Methods 4:18. https://doi.org/10.1186/1746-4811-4-18
Karlo M, Boschiero C, Landerslev KG, Blanco GS, Wen J, Mysore KS, Dai X, Zhao PX, de Bang TC (2020) The CLE53–SUNN genetic pathway negatively regulates arbuscular mycorrhiza root colonization in Medicago truncatula. J Exp Bot 71:4972–4984. https://doi.org/10.1093/jxb/eraa193
Kassaw TK, Frugoli JA (2012) Simple and efficient methods to generate split roots and grafted plants useful for long-distance signaling studies in Medicago truncatula and other small plants. Plant Methods 8:38. https://doi.org/10.1186/1746-4811-8-38
Kassaw T, Nowak S, Schnabel E, Frugoli J (2017) ROOT DETERMINED NODULATION1 is required for M. truncatula CLE12, but not CLE13, peptide signaling through the SUNN receptor kinase. Plant Physiol 174:2445–2456. https://doi.org/10.1104/pp.17.00278
Kim D, Langmead B, Salzberg SL (2015) HISAT: a fast spliced aligner with low memory requirements. Nat Methods 12:357. https://doi.org/10.1038/nmeth.3317
Knight H, Mugford SG, Ãœlker B, Gao D, Thorlby G, Knight MR (2009) Identification of SFR6, a key component in cold acclimation acting post-translationally on CBF function. Plant J 58:97–108. https://doi.org/10.1111/j.1365-313X.2008.03763.x
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Kumar A, Cousins DR, Liu C, Xu P, Murray JD (2020) Nodule inception is not required for arbuscular mycorrhizal colonization of Medicago truncatula. Plants 9:71. https://doi.org/10.3390/plants9010071
Laffont C, Ivanovici A, Gautrat P, Brault M, Djordjevic MA, Frugier F (2020) The NIN transcription factor coordinates CEP and CLE signaling peptides that regulate nodulation antagonistically. Nat Commun 11:1–13. https://doi.org/10.1038/s41467-020-16968-1
Leong SA, Williams PH, Ditta GS (1985) Analysis of the 5′ regulatory region of the gene for δ-aminolevulinic acid synthetase of Rhizobium meliloti. Nucleic Acids Res 13:5965–5976
Levy J, Bres C, Geurts R, Chalhoub B, Kulikova O, Duc G, Journet EP, Ane JM, Lauber E, Bisseling T, Denarie J, Rosenberg C, Debelle F (2004) A putative Ca2+ and calmodulin-dependent protein kinase required for bacterial and fungal symbioses. Science 303:1361–1364. https://doi.org/10.1126/science.1093038
Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, Homer N, Marth G, Abecasis G, Durbin R (2009) The sequence alignment/map format and SAMtools. Bioinformatics 25:2078–2079. https://doi.org/10.1093/bioinformatics/btp352
Limpens E, Ramos J, Franken C, Raz V, Compaan B, Franssen H, Bisseling T, Geurts R (2004) RNA interference in Agrobacterium rhizogenes-transformed roots of Arabidopsis and Medicago truncatula. J Exp Bot 55:983–992. https://doi.org/10.1093/jxb/erh122
Lin J, Li X, Luo Z, Mysore KS, Wen J, Xie F (2018) NIN interacts with NLPs to mediate nitrate inhibition of nodulation in Medicago truncatula. Nature Plants 4:942–952. https://doi.org/10.1038/s41477-018-0261-3
Liu CW, Breakspear A, Guan D, Cerri MR, Jackson K, Jiang S, Robson F, Radhakrishnan GV, Roy S, Bone C, Stacey N, Rogers C, Trick M, Niebel A, Oldroyd GED, de Carvalho-Niebel F, Murray JD (2019a) NIN Acts as a network hub controlling a growth module required for Rhizobial infection. Plant Physiol 179:1704–1722. https://doi.org/10.1104/pp.18.01572
Liu J, Rutten L, Limpens E, van der Molen T, van Velzen R, Chen R, Chen Y, Geurts R, Kohlen W, Kulikova O, Bisseling T (2019b) A remote cis-regulatory region is required for NIN expression in the pericycle to initiate nodule primordium formation in Medicago truncatula. Plant Cell 31:68–83. https://doi.org/10.1105/tpc.18.00478
Luginbuehl LH, Oldroyd GE (2017) Understanding the arbuscule at the heart of endomycorrhizal symbioses in plants. Current Biol 27:R952–R963. https://doi.org/10.1016/j.cub.2017.06.042
Marsh JF, Rakocevic A, Mitra RM, Brocard L, Sun J, Eschstruth A, Long SR, Schultze M, Ratet P, Oldroyd GE (2007) Medicago truncatula NIN is essential for rhizobial-independent nodule organogenesis induced by autoactive calcium/calmodulin-dependent protein kinase. Plant Physiol 144:324–335. https://doi.org/10.1104/pp.106.093021
Medina MJH, Gagnon H, Piché Y, Ocampo JA, Garrido JMG, Vierheilig H (2003) Root colonization by arbuscular mycorrhizal fungi is affected by the salicylic acid content of the plant. Plant Sci 164:993–998
Meixner C, Ludwig-Müller J, Miersch O, Gresshoff P, Staehelin C, Vierheilig H (2005) Lack of mycorrhizal autoregulation and phytohormonal changes in the supernodulating soybean mutant nts1007. Planta 222:709–715. https://doi.org/10.1007/s00425-005-0003-4
Mohd-Radzman NA, Laffont C, Ivanovici A, Patel N, Reid D, Stougaard J, Frugier F, Imin N, Djordjevic MA (2016) Different pathways act downstream of the CEP peptide receptor CRA2 to regulate lateral root and nodule development. Plant Physiol 171:2536–2548. https://doi.org/10.1104/pp.16.00113
Morandi D, Sagan M, Prado-Vivant E, Duc G (2000) Influence of genes determining supernodulation on root colonization by the mycorrhizal fungus glomus mosseae in Pisum sativum and Medicago truncatula mutants. Mycorrhiza 10:37–42. https://doi.org/10.1007/s005720050285
Mortier V, Den Herder G, Whitford R, Van de Velde W, Rombauts S, D’Haeseleer K, Holsters M, Goormachtig S (2010) CLE peptides control Medicago truncatula nodulation locally and systemically. Plant Physiol 153:222–237. https://doi.org/10.1104/pp.110.153718
Mortier V, De Wever E, Vuylsteke M, Holsters M, Goormachtig S (2012a) Nodule numbers are governed by interaction between CLE peptides and cytokinin signaling. Plant J 70:367–376. https://doi.org/10.1111/j.1365-3040.2011.02406.x
Mortier V, Holsters M, Goormachtig S (2012b) Never too many? How legumes control nodule numbers. Plant, Cell Environ 35:245–258. https://doi.org/10.1111/j.1365-3040.2011.02406.x
Müller LM, Flokova K, Schnabel E, Sun X, Fei Z, Frugoli J, Bouwmeester HJ, Harrison MJ (2019) A CLE–SUNN module regulates strigolactone content and fungal colonization in arbuscular mycorrhiza. Nature Plants 5:933–939. https://doi.org/10.1038/s41477-019-0501-1
Müller LM, Campos-Soriano L, Levesque-Tremblay V, Bravo A, Daniels DA, Pathak S, Park H, Harrison MJ (2020) Constitutive overexpression of RAM1 leads to an increase in arbuscule density in Brachypodium distachyon. Plant Physiol 184:1263–1272. https://doi.org/10.1104/pp.20.00997
Murray JD, Karas BJ, Sato S, Tabata S, Amyot L, Szczyglowski K (2007) A cytokinin perception mutant colonized by Rhizobium in the absence of nodule organogenesis. Science 315:101–104. https://doi.org/10.1126/science.1132514
Nagulapalli M, Maji S, Dwivedi N, Dahiya P, Thakur JK (2016) Evolution of disorder in mediator complex and its functional relevance. Nucleic Acids Res 44:1591–1612. https://doi.org/10.1093/nar/gkv1135
Oldroyd GE (2013) Speak, friend, and enter: signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol 11:252. https://doi.org/10.1038/nrmicro2990
Pecrix Y, Staton SE, Sallet E, Lelandais-Brière C, Moreau S, Carrère S, Blein T, Jardinaud M, Latrasse D, Zouine M (2018) Whole-genome landscape of Medicago truncatula symbiotic genes. Nature Plants 4:1017. https://doi.org/10.1038/s41477-018-0286-7
Penmetsa RV, Cook DR (1997) A legume ethylene-insensitive mutant hyperinfected by its Rhizobial Symbiont. Science 275:527–530. https://doi.org/10.1126/science.275.5299.527
Penmetsa RV, Frugoli JA, Smith LS, Long SR, Cook DR (2003) Dual genetic pathways controlling nodule number in Medicago truncatula. Plant Physiol 131:998–1008
Pfaffl MW (2001) A new mathematical model for relative quantification in real-time RT–PCR. Nucleic Acids Res 29:e45. https://doi.org/10.1093/nar/29.9.e45
Poehlman W, Schnabel E, Chavan S, Frugoli JA, Feltus FA (2019) Identifying temporally regulated root nodulation biomarkers using time series gene co-expression network analysis. Front Plant Sci 10:1409. https://doi.org/10.3389/fpls.2019.01409
Quandt HJ, Pühler A, Broer I (1993) Transgenic root nodules of Vicia hirsuta: a fast and efficient system for the study of gene expression in indeterminate-type nodules. MPMI-Mol Plant Microbe Interact 6:699–706
Redmond JW, Batley M, Djordjevic MA, Innes RW, Kuempel PL, Rolfe BG (1986) Flavones induce expression of nodulation genes in Rhizobium. Nature 323:632–635. https://doi.org/10.1038/323632a0
Roy S, Liu W, Nandety RS, Crook A, Mysore KS, Pislariu CI, Frugoli J, Dickstein R, Udvardi MK (2020) Celebrating 20 years of genetic discoveries in legume nodulation and symbiotic nitrogen fixation. Plant Cell 32:15–41. https://doi.org/10.1105/tpc.19.00279
Sagan M, Morandi D, Tarenghi E, Duc G (1995) Selection of nodulation and mycorrhizal mutants in the model plant Medicago truncatula (Gaertn.) after γ-ray mutagenesis. Plant Sci 111:63–71. https://doi.org/10.1016/0168-9452(95)04229-N
Schiessl K, Lilley JL, Lee T, Tamvakis I, Kohlen W, Bailey PC, Thomas A, Luptak J, Ramakrishnan K, Carpenter MD (2019) NODULE INCEPTION recruits the lateral root developmental program for symbiotic nodule organogenesis in Medicago truncatula. Current Biol 29:3657-3668.e5. https://doi.org/10.1016/j.cub.2019.09.005
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9:676–682. https://doi.org/10.1038/nmeth.2019
Schnabel E, Journet E, Carvalho-Niebel dF, Duc G, Frugoli J, (2005) The Medicago truncatula SUNN gene encodes a CLV1-like leucine-rich repeat receptor kinase that regulates nodule number and root length. Plant Mol Biol 58:809–822. https://doi.org/10.1007/s11103-005-8102-y
Schnabel E, Mukherjee A, Smith L, Kassaw T, Long S, Frugoli J (2010) The lss supernodulation mutant of Medicago truncatula reduces expression of the SUNN gene. Plant Physiol 154:1390–1402. https://doi.org/10.1104/pp.110.164889
Singh S, Katzer K, Lambert J, Cerri M, Parniske M (2014) CYCLOPS, a DNA-binding transcriptional activator, orchestrates symbiotic root nodule development. Cell Host Microbe 15:139–152. https://doi.org/10.1016/j.chom.2014.01.011
Smit P, Limpens E, Geurts R, Fedorova E, Dolgikh E, Gough C, Bisseling T (2007) Medicago LYK3, an entry receptor in rhizobial nodulation factor signaling. Plant Physiol 145:183–191. https://doi.org/10.1104/pp.107.100495
Smith SE, Read DJ (2008) Mycorrhizal symbiosis. Academic press, Cambridge
Sorek N, Szemenyei H, Sorek H, Landers A, Knight H, Bauer S, Wemmer DE, Somerville CR (2015) Identification of MEDIATOR16 as the Arabidopsis COBRA suppressor MONGOOSE1. Proc Natl Acad Sci USA 112:16048–16053. https://doi.org/10.1073/pnas.1521675112
Soutourina J (2018) Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 19:262. https://doi.org/10.1038/nrm.2017.115
Soyano T, Hirakawa H, Sato S, Hayashi M, Kawaguchi M (2014) Nodule inception creates a long-distance negative feedback loop involved in homeostatic regulation of nodule organ production. Proc Natl Acad Sci USA 111:14607–14612
Suzaki T, Takeda N, Nishida H, Hoshino M, Ito M, Misawa F, Handa Y, Miura K, Kawaguchi M (2019) LACK OF SYMBIONT ACCOMMODATION controls intracellular symbiont accommodation in root nodule and arbuscular mycorrhizal symbiosis in Lotus japonicus. PLoS Genetics 15:e1007865. https://doi.org/10.1371/journal.pgen.1007865
Tan S, Sanchez M, Laffont C, Boivin S, Le Signor C, Thompson R, Frugier F, Brault M (2020) A cytokinin signaling type-B response regulator transcription factor acting in early nodulation. Plant Physiol 183:1319–1330. https://doi.org/10.1104/pp.19.01383
Truchet G, Camut S, De Billy F, Odorico R, Vasse J (1989) The Rhizobium-legume symbiosis two methods to discriminate between nodules and other root-derived structures. Protoplasma 149:82–88
Veerappan V, Jani M, Kadel K, Troiani T, Gale R, Mayes T, Shulaev E, Wen J, Mysore KS, Azad RK (2016) Rapid identification of causative insertions underlying Medicago truncatula Tnt1 mutants defective in symbiotic nitrogen fixation from a forward genetic screen by whole genome sequencing. BMC Genom 17:1–11. https://doi.org/10.1186/s12864-016-2452-5
Vernié T, Kim J, Frances L, Ding Y, Sun J, Guan D, Niebel A, Gifford ML, de Carvalho-Niebel F, Oldroyd GE (2015) The NIN transcription factor coordinates diverse nodulation programs in different tissues of the Medicago truncatula root. Plant Cell 27:3410–3424. https://doi.org/10.1105/tpc.15.00461
Voorrips R (2002) MapChart: software for the graphical presentation of linkage maps and QTLs. J Hered 93:77–78. https://doi.org/10.1093/jhered/93.1.77
Wang C, Yao J, Du X, Zhang Y, Sun Y, Rollins JA, Mou Z (2015) The arabidopsis mediator complex subunit16 is a key component of basal resistance against the necrotrophic fungal pathogen Sclerotinia sclerotiorum. Plant Physiol 169:856–872. https://doi.org/10.1104/pp.15.00351
Wang C, Reid JB, Foo E (2018) The art of self-control–autoregulation of plant–microbe symbioses. Front Plant Sci 9:988. https://doi.org/10.3389/fpls.2018.00988
Wesly, SV, Helliwell, CA, Smith, NA, Wang, M, Rouse, DT, Liu, Q, Gooding, PS, Singh, SP, Abbott, D, Stoutjesdijk, PA, Robinson, SP, Gleave, AP, Green, AG, waterhouse, PM (2001) Construct design for efficient, effective and high-throughput gene silencing in plants, Plant J 27:581–590. https://doi.org/10.1046/j.1365-313X.2001.01105.x
Wright PE, Dyson HJ (2015) Intrinsically disordered proteins in cellular signalling and regulation. Nat Rev Mol Cell Biol 16:18–29. https://doi.org/10.1038/nrm3920
Xiao TT, Schilderink S, Moling S, Deinum EE, Kondorosi E, Franssen H, Kulikova O, Niebel A, Bisseling T (2014) Fate map of Medicago truncatula root nodules. Development. https://doi.org/10.1242/dev.110775
Xue D, Guo N, Zhang X, Zhao J, Bu Y, Jiang D, Wang X, Wang H, Guan R, Xing H (2019) Genome-wide analysis reveals the role of mediator complex in the soybean Phytophthora sojae interaction. IJMS 20:4570
Yang Y, Ou B, Zhang J, Si W, Gu H, Qin G, Qu L (2014) The arabidopsis mediator subunit MED 16 regulates iron homeostasis by associating with EIN 3/EIL 1 through subunit MED 25. Plant J 77:838–851
Zhang X, Wang C, Zhang Y, Sun Y, Mou Z (2012) The Arabidopsis mediator complex subunit16 positively regulates salicylate-mediated systemic acquired resistance and jasmonate/ethylene-induced defense pathways. Plant Cell 24:4294–4309. https://doi.org/10.1105/tpc.112.103317
Acknowledgements
This work was funded by the National Science Foundation (USA) Integrative Organismal Systems proposals # 1733470 and 1444461 to JF and USDA-NIFA 2022−67013−36881 to LMM. We acknowledge CyVerse (URL: www.cyverse.org) supported by the National Science Foundation under Award Numbers DBI-0735191, DBI-1265383, and DBI-1743442 for providing genomic sequence data storage and analysis platform. We thank Dr. Rooksana Noorai at Clemson University Genomics and Bioinformatics Facility for providing insightful suggestions on genomic data analysis. The facility is supported by Grant P20GM109094 an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health.
Author information
Authors and Affiliations
Contributions
DC, ES, AC and JF contributed to the study conception and design. Material preparation, data collection and analysis were performed by DC, ES, AC, SB, LM, and JF. The first draft of the manuscript was written by DC as part of a PhD dissertation, revised by JF and all authors commented on and contributed to versions of the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
This work was funded by NSF IOS 1733470 and 1444461 to Frugoli and USDA-NIFA 2022 − 67013 − 36881 to Müller. The authors have no relevant financial or non-financial interests to disclose.
Additional information
Handling Editor: Charitha Jayasinghege.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
344_2023_10993_MOESM3_ESM.pptx
Supplementary file3 (PPTX 709 kb)—Fig. 3 SNP index plots for the 8 chromosomes of M. truncatula in the sos1 sunn-1mutant identify a region of interest on Chromosome 4
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chaulagain, D., Schnabel, E., Crook, A. et al. A Mutation in Mediator Subunit MED16A Suppresses Nodulation and Increases Arbuscule Density in Medicago truncatula. J Plant Growth Regul 42, 7004–7022 (2023). https://doi.org/10.1007/s00344-023-10993-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-023-10993-2