Abstract
Wild sugarcane, Saccharum spontaneum, is a cold-tolerant species, and cultivated S. officinarum is highly sensitive to cold stress. The present study aimed at identifying genes and mechanisms acting in stress conditions using RNA-Seq technique to compare S. spontaneum and the cold-sensitive CP69-1062 cultivar of S. officinarum under non-stress (25 °C), chilling (0 °C), and freezing (− 4 °C) conditions. By studying 347,840,362 reads, the number of 16,088 genes showed a significant expression difference at the 1% probability level. A total of 413 genes with the highest expression changes were selected for the complementary analysis. The results showed the amount and intensity of biological activities of the cold-sensitive cultivar increased at 0 °C. By lowering the temperature to − 4 °C, the catabolic activity of the cold-sensitive cultivar drastically reduced. Reconstruction of the gene co-expression network in sugarcane showed that the LOS1 is a key gene in the chloroplast protein synthesis (elongation factor 2). Its expression profile showed that its expression level in all six samples examined was above 6000 transcript copies. It seems that the SVR3 gene plays a key role in tolerating cold stress in tolerant cultivars. Examination of the expression profile showed that with increasing cold, the SVR3 gene expression decreased in the sensitive cultivar. The expression difference of the SVR3 gene between sensitive and tolerant cultivars exceeded five times at 0 °C, and even more than 13 times at − 4 °C. Given its role in the translation process, the SVR3 gene can be considered a superiority factor of the cold-tolerant cultivar over the sensitive one. It is suggested to study the expression of LOS1 and SVR3 genes in other cold- tolerant plants for a comprehensive view of the action process of these genes. The expression levels of transcripts were validated by RT-qPCR.





Similar content being viewed by others
Notes
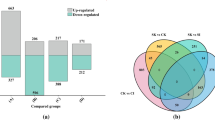
Biological process.
Molecular function.
Cellular component.
References
Apweiler R, Bairoch A, Wu CH, Barker WC, Boeckmann B, Ferro S et al (2004) UniProt: the universal protein knowledgebase. Nucleic Acids Res 32:D115–D119. https://doi.org/10.1093/nar/gkh131
Atkinson LJ, Sherlock DJ, Atkin OK (2015) Source of nitrogen associated with recovery of relative growth rate in Arabidopsis thaliana acclimated to sustained cold treatment. Plant Cell Environ 38:1023–1034. https://doi.org/10.1111/pce.12460
Bayro-Kaiser V, Nelson N (2020) Temperature sensitive photosynthesis: point mutated CEF-G, PRK, or PsbO act as temperature-controlled switches for essential photosynthetic processes. Front Plant Sci 11:562985. https://doi.org/10.3389/fpls.2020.562985
Belintani NG, Guerzoni JTS, Moreira RMP, Vieira LGE (2012) Improving low-temperature tolerance in sugarcane by expressing the ipt gene under a cold inducible promoter. Biol Plant 56:71–77. https://doi.org/10.1007/s10535-012-0018-1
Cao S, Wang Y, Li X, Gao F, Feng J, Zhou Y (2020) Characterization of the AP2/ERF transcription factor family and expression profiling of DREB subfamily under cold and osmotic stresses in Ammopiptanthus nanus. Plants (Basel, Switzerland) 9:455. https://doi.org/10.3390/plants9040455
Cronan JE Jr, Laporte D (2005) Tricarboxylic acid cycle and glyoxylate bypass. Eco Sal plus. https://doi.org/10.1128/ecosalplus.3.5.2
Ding Y, Shi Y, Yang S (2019) Advances and challenges in uncovering cold tolerance regulatory mechanisms in plants. New Phytol 222:1690–1704. https://doi.org/10.1111/nph.15696
Dong W, Ma X, Jiang H, Zhao C, Ma H (2020) Physiological and transcriptome analysis of Poa pratensis var. anceps cv. Qinghai in response to cold stress. BMC Plant Biol 20:362. https://doi.org/10.1186/s12870-020-02559-1
Engineer CB, Ghassemian M, Anderson JC, Peck SC, Hu H, Schroeder JI (2014) Carbonic anhydrases, EPF2 and a novel protease mediate CO2 control of stomatal development. Nature 513:246–250. https://doi.org/10.1038/nature13452
Gao H, Sumanaweera N, Bailer SM, Stochaj U (2003) Nuclear accumulation of the small GTPase Gsp1p depends on nucleoporins Nup133p, Rat2p/Nup120p, Nup85p, Nic96p, and the acetyl-CoA carboxylase Acc1p. J Biol Chem 278:25331–25340. https://doi.org/10.1074/jbc.m301607200
Guo L, Ma F, Wei F, Fanella B, Allen DK, Wang X (2014) Cytosolic phosphorylating glyceraldehyde-3-phosphate dehydrogenases affect Arabidopsis cellular metabolism and promote seed oil accumulation. Plant Cell 26:3023–3035. https://doi.org/10.1105/tpc.114.126946
Haldrup A, Simpson DJ, Scheller HV (2000) Down-regulation of the PSI-F subunit of photosystem I (PSI) in Arabidopsis thaliana. The PSI-F subunit is essential for photoautotrophic growth and contributes to antenna function. J Biol Chem 275:31211–31218. https://doi.org/10.1074/jbc.m002933200
Hao X, Wang B, Wang L, Zeng J, Yang Y, Wang X (2018) Comprehensive transcriptome analysis reveals common and specific genes and pathways involved in cold acclimation and cold stress in tea plant leaves. Sci Hortic 240:354–368. https://doi.org/10.1016/j.scienta.2018.06.008
Huang HL, Krieger IV, Parai MK, Gawandi VB, Sacchettini JC (2016) Mycobacterium tuberculosis malate synthase structures with fragments reveal a portal for substrate/product exchange. J Biol Chem 291:27421–27432. https://doi.org/10.1074/jbc.m116.750877
Infante C, Asensio E, Cañavate JP, Manchado M (2008) Molecular characterization and expression analysis of five different elongation factor 1 alpha genes in the flatfish Senegalese sole (Solea senegalensis Kaup): differential gene expression and thyroid hormones dependence during metamorphosis. BMC Mol Biol 9:19. https://doi.org/10.1186/1471-2199-9-19
Jia Q, Kong D, Li Q, Sun S, Song J, Zhu Y et al (2019) The function of inositol phosphatases in plant tolerance to abiotic stress. Int J Mol Sci 20:3999. https://doi.org/10.3390/ijms20163999
Kal AJ, van Zonneveld AJ, Benes V, van den Berg M, Koerkamp MG, Albermann K et al (1999) Dynamics of gene expression revealed by comparison of serial analysis of gene expression transcript profiles from yeast grown on two different carbon sources. Mol Biol Cell 10:1859–1872. https://doi.org/10.1091/mbc.10.6.1859
Livak KJ, Schmittgen TD (2001) Analysis of relative gene expression data using real-time quantitative PCR and the 2− ΔΔCT method. Methods 25:402–408. https://doi.org/10.1006/meth.2001.1262
Maere S, Heymans K, Kuiper M (2005) BiNGO: a Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21:3448–3449. https://doi.org/10.1093/bioinformatics/bti551
Mattiello L, Riaño-Pachón DM, Martins MCM, Cruz LPd, Bassi D, Marchiori PER et al (2015) Physiological and transcriptional analyses of developmental stages along sugarcane leaf. BMC Plant Biol. https://doi.org/10.1186/s12870-015-0694-z
Mekonnen T, Diro M, Sharma M, Negi T (2014) Protocol optimization for in vitro mass propagation of two sugarcane (Saccharum officinarum L.) clones grown in Ethiopia. Afr J Biotechnol. https://doi.org/10.5897/AJB2013.13575
Moliterni VM, Paris R, Onofri C, Orrù L, Cattivelli L, Pacifico D et al (2015) Early transcriptional changes in Beta vulgaris in response to low temperature. Planta 242:187–201. https://doi.org/10.1007/s00425-015-2299-z
Mousavi S, Alisoltani A, Shiran B, Fallahi H, Ebrahimie E, Imani A et al (2014) De novo transcriptome assembly and comparative analysis of differentially expressed genes in Prunus dulcis Mill. in response to freezing stress. PLoS One 9:e104541. https://doi.org/10.1371/journal.pone.0104541
Ortiz D, Hu J, Salas Fernandez MG (2017) Genetic architecture of photosynthesis in Sorghum bicolor under non-stress and cold stress conditions. J Exp Bot 68:4545–4557. https://doi.org/10.1093/jxb/erx276
Park JW, Benatti TR, Marconi T, Yu Q, Solis-Gracia N, Mora V et al (2015) Cold responsive gene expression profiling of sugarcane and Saccharum spontaneum with functional analysis of a cold inducible Saccharum homolog of NOD26-like intrinsic protein to salt and water stress. PLoS ONE 10:e0125810. https://doi.org/10.1371/journal.pone.0125810
Pearce RS (2001) Plant freezing and damage. Ann Bot 87:417–424. https://doi.org/10.1006/anbo.2000.1352
Rasheed R, Wahid A, Farooq M, Hussain I, Basra SMA (2011) Role of proline and glycinebetaine pretreatments in improving heat tolerance of sprouting sugarcane (Saccharum sp.) buds. J Plant Growth Regul 65:35–45. https://doi.org/10.1007/s10725-011-9572-3
Ream TS, Haag JR, Wierzbicki AT, Nicora CD, Norbeck AD, Zhu JK et al (2009) Subunit compositions of the RNA-silencing enzymes Pol IV and Pol V reveal their origins as specialized forms of RNA polymerase II. Mol Cell 33:192–203. https://doi.org/10.1016/j.molcel.2008.12.015
Riascos-Arcos JJ, Espitia Navarro HF, López Gerena J (2015) Assessment of the new Next Generation Sequencing (NGS) tools to identify genes associated with tolerance to water deficit in sugarcane. Acta Agron, Universidad Nacional de Colombia 64:355–362. https://doi.org/10.15446/acag.v64n4.47772
Rius SP, Casati P, Iglesias AA, Gomez-Casati DF (2008) Characterization of Arabidopsis lines deficient in GAPC-1, a cytosolic NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. Plant Physiol 148:1655–1667. https://doi.org/10.1104/pp.108.128769
Saini G, Meskauskiene R, Pijacka W, Roszak P, Sjögren LL, Clarke AK et al (2011) “Happy on norflurazon” (hon) mutations implicate perturbance of plastid homeostasis with activating stress acclimatization and changing nuclear gene expression in norflurazon-treated seedlings. Plant J 65:690–702. https://doi.org/10.1111/j.1365-313x.2010.04454.x
Saleem M, Fariduddin Q, Janda T (2020) Multifaceted role of salicylic acid in combating cold stress in plants: a review. J Plant Growth Regul. https://doi.org/10.1007/s00344-020-10152-x
Selvarajan D, Mohan C, Dhandapani V, Nerkar G, Jayanarayanan AN, Vadakkancherry Mohanan M et al (2018) Differential gene expression profiling through transcriptome approach of Saccharum spontaneum L. under low temperature stress reveals genes potentially involved in cold acclimation. 3 Biotech 8:195. https://doi.org/10.1007/s13205-018-1194-2
Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D et al (2013) Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res 13:2498–2504. https://doi.org/10.1101/gr.1239303
Shewfelt RL (1992) Response of plant membranes to chilling and freezing. plant membranes. Springer, Dordrecht, pp 192–219. https://doi.org/10.1007/978-94-017-2683-2_10
Shi YL, Cai ZY, Li D, Lu JL, Ye JH, Liang YR et al (2019) Effect of freezing on photosystem II and assessment of freezing tolerance of tea cultivar. Plants 8:434. https://doi.org/10.3390/plants8100434
Shiri Y, Solouki M, Ebrahimie E, Emamjomeh A, Zahiri J (2018) Unraveling the transcriptional complexity of compactness in sistan grape cluster. Plant Sci 270:198–208. https://doi.org/10.1016/j.plantsci.2018.02.011
Shiri Y, Khodavirdipour A, Kalkali N (2020a) Re-construction of co-expression network of genes involved in bacterial cell wall synthesis and their role in penicillin resistance. Avicenna J Clin Microbiol Infect 7:65–71. https://doi.org/10.34172/ajcmi.2020.15
Shiri Y, Solouki M, Ebrahimie E, Emamjomeh A, Zahiri J (2020b) Gibberellin causes wide transcriptional modifications in the early stage of grape cluster development. Genomics 112:820–830. https://doi.org/10.1016/j.ygeno.2019.05.022
Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M et al (2017) The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res 45(Database issue):D362–D368. https://doi.org/10.1093/nar/gkw937
Tan WJ, Yang YC, Zhou Y, Huang LP, Xu L, Chen QF et al (2018) Diacylglycerol acyltransferase and diacylglycerol kinase modulate triacylglycerol and phosphatidic acid production in the plant response to freezing stress. Plant Physiol 177:1303–1318. https://doi.org/10.1104/pp.18.00402
Tolosa LN, Zhang Z (2020) The role of major transcription factors in solanaceous food crops under different stress conditions: current and future perspectives. Plants (Basel, Switzerland). https://doi.org/10.3390/plants9010056
Untergasser A, Nijveen H, Rao X, Bisseling T, Geurts R, Leunissen JAM (2007) Primer3Plus, an enhanced web interface to Primer 3. Nucleic Acids Res 35(Web server issue):71–74. https://doi.org/10.1093/nar/gkm306
Vicentini R, Bottcher A, Dos Santos BM, Dos Santos AB, Creste S, de Andrade Landell MG et al (2015) Correction: large-scale transcriptome analysis of two sugarcane genotypes contrasting for lignin content. PloS One 10:137698. https://doi.org/10.1371/journal.pone.0137698
Wang M, Zhang X, Liu JH (2015a) Deep sequencing-based characterization of transcriptome of trifoliate orange (Poncirus trifoliata (L.) Raf.) in response to cold stress. BMC Genomics 16:555. https://doi.org/10.1186/s12864-015-1629-7
Wang X, Wei Y, Shi L, Ma X, Theg SM (2015b) New isoforms and assembly of glutamine synthetase in the leaf of wheat (Triticum aestivum L.). J Exp Bot 66:6827–6834. https://doi.org/10.1093/jxb/erv388
Wu Q, Xu L, Guo J, Su Y, Que Y (2013) Transcriptome profile analysis of sugarcane responses to Sporisorium scitaminea infection using solexa sequencing technology. Biomed Res Int 2013:298920. https://doi.org/10.1155/2013/298920
Yadav SK (2010) Cold stress tolerance mechanisms in plants. A review. Agron Sustain Dev 30:515–527. https://doi.org/10.1051/agro/2009050
Yang H, Wang T, Yu X, Yang Y, Wang C, Yang Q et al (2020) Enhanced sugar accumulation and regulated plant hormone signalling genes contribute to cold tolerance in hypoploid Saccharum spontaneum. BMC Genomics 21:507. https://doi.org/10.1186/s12864-020-06917-z
Acknowledgements
We are very grateful to the Sugarcane Research and Training Institute (Tissue Culture Laboratory and Research Greenhouse) related to Khuzestan Sugarcane and By-products Development Company, for helping us in this project.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial or not-for-profit sectors.
Author information
Authors and Affiliations
Contributions
MN, and BAF, conceived and designed the manuscript, wrote the manuscript and contributed equally. EE performed the experiments. AE performed the experiments and edited the manuscript. JZ performed the experiments, analyzed the data. MPA performed the experiments.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Handling Editor: Fabrizio Costa.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Neisi, M., Fakheri, B.A., Ebrahimie, E. et al. RNA-Seq Analyzing Reveals the Key Role of SVR3 Gene in Tolerating the Freezing Stress in Saccharum spontaneum. J Plant Growth Regul 42, 2613–2625 (2023). https://doi.org/10.1007/s00344-022-10731-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-022-10731-0