Abstract
Gibberellin (GA), auxin (IAA) and brassinosteroid (BR) are indispensable in the process of plant growth and development. Currently, research on the regulatory mechanism of phytohormones in banana dwarfism is mainly focused on GA, and few studies are focused on IAA and BR. In this study, we measured the contents of endogenous GA, IAA and BR and compared the transcriptomes of wild-type Williams banana and its dwarf mutant across five successive growth periods. We investigated the relationship between hormones and banana dwarfism and explored differential gene expression through transcriptome analysis, thus revealing the possible metabolic regulatory mechanism. We inferred a complex regulatory network of banana dwarfing. In terms of endogenous hormone levels, GA and IAA had significant effects on banana dwarfing, while BR had little effect. The key gene in GA biosynthesis of is GA2ox, and the key genes in IAA biosynthesis are TDC and YUCCA. The differential expression of these genes might be the main factor affecting hormone levels and plant height. In terms of hormone signal transduction, DELLA and AUX/IAA repressor proteins were the core regulators of GA and IAA, respectively. They inhibited the process of signal transduction and had feedback regulation on hormone levels. Finally, the transporter protein PIN, AUX1/LAX protein family and ABCB subfamily played supplementary roles in the transport of IAA. These results provide new insights into GA and IAA regulation of banana growth and a reliable foundation for the improvement of dwarf varieties.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana (plantain) is one of the most important fruit crops worldwide and is of great significance to human life. It is not only a popular tropical high-quality fruit but also an important staple crop for more than 400 million people in the tropics. Generally, the height of cultivated banana is more than 2 m, which makes the plant prone to lodging under the influence of typhoons and storms, resulting in great economic loss.
Cultivation of dwarf plants has always been one of the important objectives of modern breeding. Since the last century, a ‘Green revolution’ in crop breeding has been initiated, along with the discovery of plant dwarfing genes and the international selection of dwarfing varieties of rice and wheat (Hedden 2003). Through breeding for dwarf varieties, scientists have obtained many ‘modern varieties’ of crops with dwarfing characteristics. However, banana has remained practically unaffected under the ‘Green revolution’ due to its characteristic triploid sterility, which limits conventional breeding (Dash and Rai 2016).
Currently, the phenotype and mechanism of banana dwarfing have not been thoroughly studied because of the lack of dwarfing materials available for study. The reported studies on banana dwarfing mainly focused on dwarf mutant materials, including the analysis and identification of dwarf mutant plants in tissue culture and mutation generation using RAPD (Damasco et al. 1996), AFLP and SCAR.
It has been reported that mutants with defects in plant hormone biosynthesis or signaling could result in dwarfism (Chen et al. 2016; Durbak et al. 2012; Richards et al. 2001; Wei et al. 2018). Gibberellin (GA), auxin (IAA) and brassinosteroid (BR) are three important phytohormones well known to control and regulate plant growth and development (Ma et al. 2016; Nemhauser et al. 2006; Wang et al. 2018). The GAs content was significantly different between Williams banana and its dwarf mutant, as well as between those lines and the dwarfing type recovered under exogenous GA3 application. A series of subsequent studies were carried out to further explore the dwarfing mechanisms and discover dwarfing genes. Genome-wide screening revealed 36 candidate GA metabolism genes, which were systematically identified for the first time (Chen et al. 2016). Shao illustrated the effect of GA on banana dwarfing. They obtained six mutant lines through gene editing of five GA20ox2 homologous genes in the banana cultivar ‘Gros Michel’ using the CRISPR/Cas9 system. The height of the mutants was significantly reduced compared with that of the wild type at three developmental stages (Shao et al. 2020).
At present, most of the research on the effects of plant hormones on banana dwarfism has mainly focused on the regulatory mechanism of GA, but there is limited knowledge about the effects of IAA and BR. In this study, we analyzed the changes in the contents of GAs, IAA and BR between wild‐type Williams banana and its dwarf mutant to reveal the possible relationship between plant hormone changes and banana dwarfism.
Nanning has a subtropical monsoon climate, with an average annual temperature of 21.6 degrees and an average annual rainfall of 1304.2 mm. The lowest temperature is in December, with an average of 12.8 degrees. The highest temperature is in July and August, with an average of 28.2 degrees. The suitable temperature and sufficient water make Nanning one of the major banana-producing areas. In this work, WGH and ‘WGH Dwarf’ were taken as the research group. ‘Tianbao’ banana and ‘Nalong Dwarf’ banana were used as the reference group. We measured the contents of endogenous GA, IAA and BR across five successive growth periods and performed transcriptome analysis. The correlation analysis that combined the hormone level results and plant heights was performed to reveal the effects of endogenous GAs, IAA and BR on plant height. Then, we proposed a possible explanation for banana dwarfism, hoping to contribute to the following research on banana dwarfism breeding.
Materials and Methods
Plant Materials and Cultivation Conditions
In this work, four banana resources were selected for experimentation. ‘WGH Dwarf’ (referred to as W2) is a stable dwarf mutant of wild-type Williams banana ‘WGH’ (Musa spp. AAA group) (referred to as H2). In addition, the comparison between ‘Tianbao’ banana (Musa spp. AAA group, referred to as H1) and ‘Nalong Dwarf’ banana (Musa spp. AAA group, referred to as W1) was selected as reference. All the materials used in this research were grown in the same field in Nanning, China, with the same growing environment and the same standard cultivation mode of fertilizer and water management. Each banana resource had 3 biological repeats. The second generation of sucker tissues without any diseases or insect damage was selected for experimentation, and the growth period of all these plants was basically the same when sampled and measured. The pseudostem heights were recorded on April 11, April 25, May 13, May 28 and July 1, which relatively evenly covered the vegetative growth period before the budding stage of banana plants. During this period, the temperature and other conditions were suitable for growth, 1–2 new leaves sprouted every 15 days, and the pseudostem gradually elongated. Moreover, the first leaf tip of the plants was selected for sampling at every time point. At 10–11 o'clock am at each sampling timepoint, 5 ~ 8 cm of the tip was taken and torn in half along the vein. One half was quickly frozen with liquid nitrogen for RNA extraction, and the other half was stored in dry ice for hormone detection.
Detection of Plant Hormones
Enzyme-linked immunosorbent assay (ELISA) was used for the detection of plant hormones (GA1, GA3, GA4, BR, and IAA). The kit was produced by Jianglai Biological Co., Ltd. (Shanghai, China) (Cat.no. JL47815, JL22720, JL47820, JL14098, JL22806). The experimental procedure followed the instructions of the kit. Each tissue sample was repeated twice for the analysis of hormone content, and 0.1 g fresh weight of tissue was taken each time.
Data Processing and RNA-seq Analysis
A total of 60 samples (4 resources * 5 time points * 3 biological repeats) were prepared and analyzed by Genedenovo Biotechnology Co., Ltd. (Guangzhou, China), including RNA extraction, construction of cDNA libraries, quality detection of RNA and cRNA libraries, RNA-seq and transcriptome analysis.
The RNA-seq libraries were sequenced on the Illumina sequencing platform. All data in this study can be viewed at the National Center for Biotechnology Information (NCBI) under the bioproject number PRJNA631124. First, we used fastp (version 0.18.0) to filter low-quality data and obtain clean reads. Then, the short read alignment tool Bowtie2 (version 2.2.8) was used to map reads to the ribosomal RNA (rRNA) database. The rRNA mapped reads were then removed. The remaining clean reads were further used for assembly and gene abundance calculations. The paired-end clean reads were mapped to the reference genome using HISAT2.2.4. The reference genome data can be downloaded from https://www.ncbi.nlm.nih.gov/assembly/GCF_000313855.2/.
The mapped reads of each sample were assembled through StringTie v1.3.1, and an FPKM (fragment per kilobase of transcript per million mapped reads) value was calculated to quantify expression abundance and variation. Then, the genes discovered in the sequencing results were aligned with the genes included in the reference genome. The genes that matched the genes in the reference genome were called “Refer Genes”, while the others were called “Novel genes”.
Screening and Functional Analysis of Differentially Expressed Genes
The differential expression of genes was analyzed by DESeq2. Our imported data included the read counts obtained from the gene expression level analysis, and the genes that met the selection criterion of P < 0.05 and |log2FC|> 1 were regarded as significantly differentially expressed genes (DEGs). All the DEGs were submitted to GO annotation term (http://www.geneontology.org/) and KEGG pathway enrichment (http://www.genome.jp/kegg/) analysis for functional annotation, and the genes with Bonferroni (Q-value) < 0.05 were regarded as significant. Using the online analysis platform OmicShare (www.omicshare.com), we screened out the genes related to the biosynthesis and signaling of GA, BR and IAA from the KEGG pathway; we show the expression differences and changes in the KEGG pathway in a schematic sketch.
WGCNA
WGCNA (weighted gene coexpression network analysis) is an analysis method for the gene expression patterns of multiple samples. It can cluster genes with similar expression patterns and analyze the correlation between each module and specific traits or phenotypes. Coexpression networks were constructed using the WGCNA (v1.47) package in R (Langfelder and Horvath 2008). After filtering the genes with low expression, gene expression values were imported into WGCNA to construct coexpression modules using the automatic network construction function blockwise modules with default settings, except that the power was 15, TOM type was unsigned, merge cut height was 0.8, and min module size was 50.
To identify significant modules, module eigengenes were used to calculate the correlation coefficient data for specific traits. Pearson’s correlation between each gene and trait within each module was also calculated for the most relevant modules (positive correlation and negative correlation) corresponding to each phenotype, and the gene significance value (GS) was obtained.
qRT-PCR Analysis
To verify the RNA-seq data, eight genes were selected for qRT-PCR analysis. The three biological repeats of each material at the same time point were mixed as one sample. Total RNA was extracted from all 20 samples (4 resources * 5 time points), and cDNA was obtained by a reverse transcription kit (R223, Vazyme Biotech, Nanjing, China). ABI Step One Plus (ABI, USA) was used to carry out fluorescence quantitative PCR, and each sample was repeated three times. The reaction system included 5 μl of 2 × ChamQ SYBR qPCR Master Mix (Q341, Vazyme Biotech, Nanjing, China), 0.4 μl of 0.01 mM dissolved forward and reverse primer (Table S2) and 5 μl of cDNA. The final system had a volume of 20 μl. The cycling conditions were as follows: initial denaturation for 90 s at 95 °C, followed by 40 cycles of 5 s at 95 °C, 15 s at 60 °C, and 20 s at 72 °C. UBQ10 (ncbi_103994099) was chosen as the reference gene. The data were processed according to the Ct (2−ΔΔCt) method.
Statistical Analysis
All statistics and data for plant heights, hormone contents and qRT-PCR were processed by Excel 365. The significance of the data was calculated by one-way ANOVA in Excel 365. Under the criteria of F > F crit, a P-value < 0.05 was regarded as significantly different, and a P-value < 0.01 was regarded as extremely significantly different. The figures, pathway diagrams and flow charts were drawn and corrected by Adobe Illustrator CC 2017.
Results
Analysis of Plant Height Differences and Hormone Content Changes
‘Tianbao’ banana (Musa spp. AAA group, referred to as H1), ‘Nalong Dwarf’ banana (Musa spp. AAA group, referred to as W1) and Williams banana ‘WGH’ (Musa spp. AAA group, referred to as H2) are three elite, high-quality banana varieties. In a previous study by our team, the stable dwarf mutant ‘WGH Dwarf’ (referred to as W2) was obtained from bud mutations by continuous tissue culture. In this work, we take H2 and W2 as the research group, where H2 is used as a control. H1 and W1 were selected for the comparison of plant height, where H1 was used as a control. There was a significant difference in the pseudostem height at flowering between the two groups (W1 vs. H1, W2 vs. H2, P < 0.01, Fig. 1A). In addition, the morphological characteristics of W2 were also significantly different from those of H2, with a smaller leaf spacing. Table 1 describes the general characteristics of these four materials over three consecutive years (summer of 2017–2019).
A The growth of plants at the bud stage. Bar = 1 m; B Plant heights at 5 time points. The line connects the changes in height in the same period. C The relationship between hormone concentration and plant height. Red represents a positive correlation; blue represents a negative correlation. In each grid, the upper numbers represent the Pearson correlation coefficient between plant height and hormone concentration, and the lower numbers represent the corresponding P value (the test value of the correlation between the two modules, calculated by Student’s t test). D–H Mean values of hormone contents. The abscissa represents different growth periods, and the ordinate represents hormone contents. The five small squares represent the significant differences at the five time points. The asterisks behind the squares represent the significance of the whole period (excluding the fourth time point). *P < 0.05, **P < 0.01; no mark indicates no significant difference (Color figure online)
GA1, GA3, GA4, BR and IAA contents were determined in the leaves of the two groups (W1 vs. H1, W2 vs. H2). Overall, the contents of GA1 and GA4 were lower in dwarf bananas (W1 and W2) than in the corresponding wild-type bananas (H1 and H2), while the contents of GA3, BR and IAA were higher in the dwarf bananas. Regarding the contents of GA1, GA4 and IAA, the differences in phytohormone levels between the dwarf banana and the corresponding wild-type banana were extremely significant (P < 0.01), while the contents of BR and GA3 were not significantly different (P > 0.05, Fig. 1D–H). Similar results were described by Chen et al (2016); compared with “8818”, the GAs content in the pseudostem and root of its dwarf mutant "8818–1" was significantly lower. Interestingly, this trend was not present in young leaves. Likewise, dwarf banana "# 31" obtained by gene editing had the same level of GAs (GA1, GA3, GA4) as its wild type (Shao et al. 2020). It should be noted that the materials used in these studies were in the seedling growth stage, and they grew relatively slowly. In this study, the selected sampling time points were in the vegetative growth period of the plant. The plants grew more vigorously in the vegetative growth period, the number of leaves and leaf area increased rapidly along with stem elongation, and hormone metabolism was much more active, so the change in the hormones in leaves in this study probably had a greater correlation with plant height.
By calculating the Pearson correlation coefficient and P value of the plant heights and contents of different hormones (Fig. 1C), it can be seen that the GA4 level had the highest correlation with plant height, followed by GA1, BR and IAA, with quite high correlation coefficients and P < 0.01, while the GA3 level had a lower correlation and larger P value. This result suggested that banana dwarfism was closely related to the contents of the endogenous hormones GA1, GA4 and IAA. BR may play a small role, but GA3 is likely not involved. Notably, at the first, second, third and fifth timepoints of the experiment, the height of dwarf banana at each time point was significantly shorter than that of the corresponding wild-type banana (P < 0.05, Figs. 1B, S1). However, the increase in height at the fourth time point was much slower than expected (Fig. 1B). Additionally, the contents of GA, IAA and BR fluctuated greatly (Fig. 1D–H).
This inconsistency may be caused by the influence of continuous rainy days around the fourth timepoint, resulting in abnormal changes in plant hormones. Therefore, the experimental data at the fourth time point may not reflect the normal growth and metabolism changes of the plant, so they were temporarily removed in the subsequent analysis.
RNA-seq Analysis and Functional Analysis of DEGs
To further explore the differential expression of genes between dwarf banana plants (W1 and W2) and their controls (H1 and H2), we analyzed the RNA-seq and transcriptome data of all 60 samples from four experimental materials at five time points. A total of 36,419,962 ~ 55,357,258 clean reads without adaptor sequences, low-quality reads or N-containing reads were obtained from each library, with a clean read ratio of 99.74% ~ 99.90% and an average output of 45,621,928 reads for each sample (Fig. 2, Table S3). Other than one library with a relatively low mapping ratio (56.8%), the other libraries had mapping ratios against the reference genome that ranged from 74.80% to 91.19%. It can be concluded that the quality of the sequencing results was high, and the results were reliable. We counted the number of reference genes and novel genes that aligned in the sequencing data. Among the 32,890 genes obtained, 30,737 were reference genes, and 2153 were novel genes, with a mapping ratio between 93.47% and 93.95%. The data from each sample is shown in Fig. S2.
With the RNA-seq data, the gene expression of each dwarf banana and its control (W1 vs. H1, W2 vs. H2) were compared at each time point, and the analysis of differentially expressed genes (DEGs) was carried out. A total of 12,974 DEGs were obtained. As shown in Fig. 3, there were more upregulated genes than downregulated genes in dwarf bananas.
Remarkably, the number of DEGs at the fourth timepoint was significantly higher than that at other time points (Fig. 3). This phenomenon may be caused by unusual climate fluctuations, which resulted in an unusual change in gene expression. Since there was no significant difference in the change in the hormone contents and plant heights at the fourth time point (Fig. 1), it can be inferred that the differentially expressed genes at this time point had little effect on the change in plant heights and hormone contents. Thus, the genes that were differentially expressed solely at the 4th time point were regarded as interference. When selecting genes closely related to plant height and hormone changes, we excluded these interfering genes from the RNA-seq analysis.
After excluding the DEGs from the fourth time point, there were a total of 6438 DEGs between dwarf bananas and their controls. These DEGs were annotated via GO term and KEGG pathway analysis. GO term classification showed that 1300 DEGs were classified into three top-level ontologies and 44 secondary functional GO terms. In the biological process category, 963 DEGs were classified into 20 functional groups, among which “metabolic”, “cellular” and “single-organism” were the most abundant terms. Forty-five out of 301 genes belonged to the “signaling” term as well. In the molecular function category, the DEGs were classified into 10 secondary functional GO terms, and most of the DEGs were concentrated in the term “catalytic activity and binding”. In the cellular component category, the DEGs were classified into 14 secondary functional GO terms, and the top three terms were “cell part”, “cell” and “membrane”. The top terms in each category after removing several GO terms with little enrichment or statistical value are shown in Fig. 4.
The KEGG pathway enrichment results showed that 1444 DEGs were classified into 128 KEGG pathways. The top 20 statistically significant enriched pathways based on the difference in the significance values of genes enriched in each pathway (P value) are shown in Fig. 5. The top 3 pathways were “plant hormone signal transduction”, “biosynthesis of secondary metabolites” and “MAPK signaling pathway-plant”. The metabolic pathways we mainly focused on were those related to the biosynthesis of GA, BR and IAA, including “monoterpenoid biosynthesis”, “sesquiterpenoid and triterpenoid biosynthesis”, “zeatin biosynthesis”, “tropine”, “piperidine and pyridine alkaloid biosynthesis” and “brassinosteroid biosynthesis”. Additionally, the “plant hormone signal transduction” pathway had quite a high enrichment degree and was significant (P < 0.05). These pathways may be significantly related to the dwarf phenotype. In addition, some other pathways, such as “biosynthesis of secondary metabolites”, “plant-pathogen interaction”, “MAPK signaling pathway-plant”, “phenylpropanoid biosynthesis”, and “flavor biosynthesis”, were found to be enriched, indicating that they may have a role in dwarfism.
Bubble diagram of the top 20 enriched KEGG pathway classifications of the identified DEGs. The abscissa represents the rich ratio, which is the ratio of the number of genes annotated to a particular item to the total number of genes in this item in one species. The calculation formula was Rich Ratio = Term Candidate Gene Num/Term Gene Num. The ordinate represents the KEGG pathway. The size of the bubbles indicates the number of genes annotated to a KEGG pathway, and the color represents the Q-value. The deeper the color is, the smaller the Q-value (Color figure online)
Analysis of DEGs Involved in GA, IAA and BR Biosynthesis
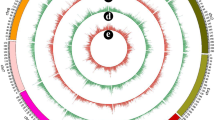
Within the 6438 selected DEGs, KEGG pathway analysis of the DEGs related to the biosynthesis of GA, IAA and BR was also carried out. First, we selected 6 DEGs related to GA biosynthesis in ko00904, 12 DEGs related to BR biosynthesis in ko00905, and 9 DEGs related to IAA biosynthesis in ko00380. Second, we ruled out the DEGs with low levels of expression in these pathways (4 YUCCA genes, 1 GA3ox gene, 1 GA2ox gene, 2 DWF4 genes and 1 BAS1 gene), and only 19 DEGs remained. After normalizing their expression levels, we classified these 19 DEGs and produced a heat map of the remaining 19 DEGs. Figure 6 shows a schematic diagram of the differentially expressed genes and their roles in the biosynthesis pathway.
Schematic diagram of GA (A), IAA (B) and BR (C) biosynthesis. Heat map representing the expression dynamics of genes associated with hormone metabolism at five timepoints. Red represents upregulation, and green represents downregulation. 1–1 to 1–5 represent the 5 timepoints of W1-H1, and 2–1 to 2–5 represent the 5 timepoints of W2-H2 (Color figure online)
In the GA biosynthesis pathway, DEGs were not significantly different at most time points. However, across all timepoints, GA3ox genes were upregulated at the early stage and then downregulated at the late stage in dwarf banana plants (W1 and W2). GA2ox genes were downregulated at each timepoint in both dwarf banana plants (W1 and W2) and their corresponding control plants (H1 and H2).
In the IAA biosynthesis pathway, during almost the whole growth process, the TDC and YUCCA genes were upregulated in W1 but downregulated in W2. The AAO gene was upregulated drastically during the whole period of growth.
In the BR biosynthesis pathway, the DET2 and DWF4/D11 genes were upregulated at the early stage and then downregulated at the late stage in dwarf banana plants. The ROT3 and CYP92A6 genes were downregulated, while the D2, BR6ox1,2 and BSA1 genes were upregulated at each time point.
Overall, the enrichment degree of DEGs in the three KEGG pathways was not high, and the relationship between gene expression and hormone content changes did not match the results of published papers. For instance, the upregulation of GA3ox genes or downregulation of GA2ox genes caused an increase in GA1 and GA4 contents (Yamaguchi 2008). However, in our research, these two hormone contents were lower in dwarf banana plants, which was contrary to the expected results. Similar contradictions were also found in the biosynthesis pathways of IAA and BR. These phenomena suggested that the mechanisms of hormone metabolism regulation were not the same among different plant species, which may be much more complex than we expected. There was no significant difference in the expression levels of genes related to the metabolic pathways of GA (P = 0.15 in ko00904) and IAA (P = 1 in ko00380). Although the DEGs in the metabolic pathway of BR changed significantly (P = 0.003 in ko00905), there was no significant difference in the hormone level.
We also noted that most of the coding genes had the same up- or downregulation trend in groups W1-H1 and W2-H2. The increase or decrease in hormone levels (Fig. 1) was consistent with the up- or downregulated expression levels of their synthetic genes at every timepoint, especially the first three timepoints. The differential expression of many genes was positively or negatively related to changes in hormone contents. It can be concluded that the differential expression of genes in metabolic pathways can affect the change in the content of the corresponding hormone.
Analysis of DEGs Involved in GA, IAA and BR Signal Transduction
There were 147 DEGs out of 481 genes in the plant hormone signal transduction pathway (ko04075), and the proportion of DEGs was 0.31. The plant hormone signal transduction pathway ranked first according to the P value (-log10(P value) = 8.94), demonstrating an extremely significant difference and indicating that it was closely related to dwarfing in banana plants. In this pathway, we found 6, 11 and 40 DEGs related to the signal transduction of GA, BR and IAA, respectively. A schematic diagram of the hormone signal transduction and expression differences of related genes is shown in Fig. 7.
Figure 7 shows the differential expression of genes related to plant hormone signal transduction between dwarf banana and the corresponding control. The DEGs in the GA signaling pathway, such as GID1 (GID1C), DELLA (GAI, RHT1) and TF (PIF4, PIL13, PIL15), were significantly upregulated at almost every time point in dwarf banana. In the BR signaling pathway, BRI1 and BKI1 were upregulated, while TCH4 and CYCD3 were downregulated in dwarf banana. However, IAA signal transduction was more complex, and many coding genes showed irregular changes. For instance, all the DEGs were upregulated in W1 during the whole growth process. However, in W2, they were upregulated at some time points and downregulated at others. It was necessary to further explore and analyze the major genes related to the regulation of plant height and hormones in dwarf mutant banana.
Using WGCNA to Analyze the DEGs Related to Hormone Levels and Plant Height
Many DEGs were identified in the anabolism and signal transduction pathways of GA, IAA and BR. However, the up- or downregulation of many DEGs, such as YUCCA, ALDG or AUX/IAA in the IAA biosynthesis pathway, was different across different groups or different time points. In this complex case of differential gene expression, we used the WGCNA method to further search for genes with a high correlation with plant height and hormone levels.
First, a hierarchical clustering tree was constructed according to the correlation between gene expression levels. Then, according to the clustering relationship between genes, the DEGs were divided into 21 modules. Each module contained genes with a similar expression pattern. Each module is represented by a different color, and the gray color represents the genes that could be classified into any module (Fig. 8A). To determine the genes related to characteristics in the modules, correlation analyses were carried out between the eigenvalues of each module and the hormone level and plant height data to calculate the Pearson correlation coefficient and significance value between each module and trait (Fig. 8B). Based on this method, we finally obtained the gene significance value (GS), which is an important analysis parameter of hormone metabolism and plant height (Fig. S3).
WGCNA of the DEGs related to hormone levels and plant height. A Gene dendrogram after dynamic tree cutting and after dynamic merging. B Correlation of each module identified by WGCNA to hormones and height. The upper number in each small square represents the Pearson correlation coefficient, and the number in parentheses represents the significance (P-value) of each module compared to each external factor. The depth of each color corresponds to the depth of correlation, with a positive correlation indicated in red and a negative correlation indicated in green. The smaller the P-value is, the stronger the significance (Color figure online)
We screened several modules with a high correlation with hormone biosynthesis and signaling from the WGCNA results (P < 0.5). Concerning GA1, 4 modules were selected: “lavenderblush3”, “darkseagreen4”, “coral2” and “cyan”. Concerning GA3, 4 modules were selected: “darkseagreen4”, “lavenderblush3”, “mediumorchid” and “brown2”. Concerning GA4, 2 modules were selected: “coral2” and “darkviolet”. Concerning IAA, 5 modules were selected: “lightsteelblue1”, “honeydew1”, “mediumorchid”, “brown” and “lightcyan”. Concerning BR, 4 modules were selected: “coral2”, “lavenderblush3”, “brown” and “cyan”. The selected modules were combined into a new module for each of the five hormones, named “WGCNA-GA1”, “WGCNA-GA3”, “WGCNA-GA4”, “WGCNA-IAA” and “WGCNA-BR”. It can be concluded that the genes in each new module had the highest correlation with the corresponding hormone contents.
Then, we analyzed the DEGs in each new module to identify the key genes in the corresponding hormone biosynthesis and signal transduction processes. The number of genes selected is shown in Venn diagrams (Fig. S4). The results further showed that there were 2 genes that had a high correlation with GAs contents: GAI (ncbi_103990128) and GID1C (ncbi_104000617). There were 4 genes that had a high correlation with IAA contents: TYDC2 (ncbi_103990339), YUC11 (ncbi_103994209), IAA6 (ncbi_103978455) and IAA27 (ncbi_103991365). However, no DEGs with a high correlation with BR contents were found.
Using the same method, we selected five modules significantly related to plant height: "indianred 4", "darkviolet", " lavendrblush3", "honeydew 1" and "brown 2". Then, the genes in these modules were combined into a new gene set (WGCNA-Height). All genes in this gene set were regarded as significantly related to plant height in the subsequent analysis. The KEGG pathway analysis showed that 176 DEGs were involved in 96 pathways. According to the enrichment results, the top 10 KEGG pathways are shown in Fig. 9. The DEGs were mainly enriched in “metabolic pathways” and “biosynthesis of secondary metabolites”. Other pathways, including “phenylpropanoid biosynthesis”, “fructose and mannose metabolism”, “amino sugar and nucleotide sugar metabolism” and “plant-pathogen interaction”, were found to be enriched.
We also noticed that in the KEGG enrichment graphic of the gene set with a high correlation with plant height, “plant hormone signal transformation” had a relatively high enrichment degree. There were 10 DEGs, 6 of which were in the IAA, GA and BR signal transduction pathways, including 4 coding genes in the IAA pathway, AUX22D (ncbi_103995122), AUX22E (ncbi_103983123), IAA30 (ncbi_103984124) and SAUR32 (ncbi_103971028). In addition, GAI (ncbi_103990128) in the GA pathway and CURL3 (ncbi_103973700) in the BR pathway were also present. Additionally, in the KEGG functional enrichment analysis of the WGCNA_Height gene set, several DEGs were related to hormone biosynthesis and signal transduction, such as GA2ox8 (ncbi_103997678) in the GA anabolic pathway, CYP75B2 (ncbi_103975053) in the BR anabolic pathway, and TYDC2 (ncbi_103990399), TDC1 (ncbi_103999582) and YUC11 (ncbi_103994209) in the IAA anabolic pathway. These genes were supposed to be more closely related to plant height.
Verification of DEGs by qRT-PCR
The expression level of eight genes related to hormone biosynthesis and signal transduction (Table S2) was verified by qRT-PCR. In the 80 verifications (8 genes * 2 groups * 5 time points), the relative expression level of 62 comparison groups was consistent with that of RNA-seq, and the compliance rate reached 77.5% (Fig. 10). The inconsistent results were mainly concentrated within the time points where the gene expression differences were not significant. The overall correlation coefficient of the linear regression analysis was 0.9126. This result suggested that our RNA-seq data and analyses were robust.
Verification of DEGs by qRT-PCR. A–H Transcript levels of 8 genes related to hormone biosynthesis and signal transduction. Data are the means of three replicates, and error bars represent ± SE (n = 3). I Pearson’s correlation of gene expression ratios between RNA-seq and qRT-PCR results. The correlation of the fold change between RNA-seq (abscissa) and qRT-PCR (ordinate) data was analyzed (Color figure online)
Discussion
Some studies have shown that the distribution of hormones in plants is uneven, and the hormone levels in roots, stems, leaves and other parts of plants vary (Clouse and Sasse 2018; Shao et al. 2020). In this study, it was necessary to analyze the same plant continuously from the seedling stage. Due to this special limitation, we sampled leaves rather than pseudostems to reduce the effect on the growth of plants. The results showed that there were significant differences in the contents of GA1, GA4 and IAA in the leaves of dwarf banana and its corresponding control, which confirmed the reliability and rationality of the sampling method. In this study, there were significant differences in plant height and in the hormone contents in leaves between dwarf banana and the control materials. In addition, according to the RNA-seq analysis, there were significant differences in DEGs related to the biosynthesis and signaling of many hormones, such as GA, BR and IAA (Table S1). Considering that the differences in hormone contents and plant heights were caused by the differences in gene expression, we tried to look for the relationship between dwarfism and the expression pattern of these DEGs to understand the molecular mechanism of hormones affecting plant growth.
The Relationship Between GA and Banana Dwarfism
Bioactive GAs mainly include GA1, GA3, GA4, and GA7 (Yamaguchi 2008). Among them, GA1 and GA4 have been confirmed to play an essential role in plant dwarfism in many species (MacMillan 2001) and are known as the main growth-active GAs (Yamaguchi 2008). In this study, the contents of GA1 and GA4 in dwarf banana plants were significantly lower than those in the corresponding control, which showed a significant correlation with plant dwarfing (P < 0.05). The lack of bioactive GAs can cause dwarfism in many species, including banana, which can be restored by the exogenous application of bioactive GAs (Chen et al. 2016; Richards et al. 2001; Shao et al. 2020). It is well known that dynamic changes in the GA content and metabolism directly affect the height of plants, but the specific mechanism in banana is still unknown. In this study, we aimed to explore the differentially expressed genes that cause variation in dwarfism in banana based on hormone metabolism and signal transduction processes.
Some reports have shown that the 2OG-Fe(II) oxygenase superfamily is involved in the mid-late stage catalysis of the synthesis of GA, among which GA20ox, GA3ox and GA2ox play crucial catalytic roles (Han and Zhu 2011). These genes affected the contents of both bioactive GA1 and GA4. The expression of GA20ox and GA3ox can increase the level of active GAs in plants. In contrast, the expression of GA2ox repressed the production of active GAs (Hedden and Kamiya 1997; Sun and Gubler 2004). Plants with loss-of-function mutations in GA20ox and GA3ox displayed a dwarf phenotype. On the other hand, overexpression of the GA2ox gene also produced dwarf plants (Ma et al. 2016). In our study, it was unexpected that the expression of GA20ox in the dwarf plants did not show a significant difference compared to that in the control materials, the expression of GA3ox was downregulated at the early stage and upregulated at the later stage, and GA2ox was significantly downregulated. According to the inference in the published reports, it can be assumed that the differential expression of GA20ox, GA3ox and GA2ox we detected could cause higher levels of GA1 and GA4 in dwarf bananas; this result was inconsistent with those of the GA1 and GA4 content in this study (Fig. 1F, H). This paradox may be due to the feedback regulation of the expression of these enzymes and suggests that there are more complex signal transduction mechanisms in the physiological regulation mechanism of banana dwarfing.
The gene expression of enzymes involved in the later stage of GA biosynthesis is usually regulated by GA content and environmental signals (such as light) (Hedden and Kamiya 1997). The regulation of the GA content is mainly because the content of active GA1 and GA4 negatively regulates GA20ox and GA3ox and positively regulates GA2ox (Olszewski et al. 2002; Thomas et al. 1999). This regulation is called feedback regulation. Negative feedback regulation of GA20ox and GA3ox at the mRNA level has been demonstrated in several plant species (Hedden and Kamiya 1997; Yamaguchi 2008). Phillips found that the transcriptional levels of three GA20ox genes were higher in the ga1-3 (CPS deficient type) mutant than in Landsberg erecta in Arabidopsis, and their transcriptional levels were greatly reduced in the mutant type after exogenous GA application (Phillips et al. 2002). Reduced GA levels caused subsequent signal transduction, resulting in the increased transcription of AtGA3ox1 in Arabidopsis, while increased GA levels caused the opposite effect (Dill and Sun 2001). The levels of GA4 and Le were upregulated in GA-deficient mutants and downregulated after GA application, showing feedback inhibition (Martin et al. 1997).
In bananas with extremely significant dwarfism, the concentrations of active GAs (GA1 and GA4) were relatively low at the early growth stage (Fig. 1F, H). As an influential feedback loop, the expression of GA3ox was downregulated at the early stage and then upregulated, while GA2ox was not significantly different at the early stage and then downregulated.
The results of the hormone test showed that the contents of GA1 and GA4 were significantly lower in the leaves of dwarf banana than in those of the control material. However, the difference in GA3 content was not significant between dwarf banana and control materials, and the GA3 content in dwarf banana was even higher than that in the control material in both groups. The change trend of the GA3 content may not be large enough to have an obvious impact on plant height, so its role in plant height may not be significant. This result was similar to that of Chen and Shao (Chen et al. 2016; Shao et al. 2020). In addition, exogenous active GAs (including GA1, GA3, GA4, and GA7) can reduce negative feedback regulation and directly participate in metabolism and signal transduction so that plant height can be restored (Chiang et al. 1995; Phillips et al. 1995; Richards et al. 2001).
There is a close interaction between metabolism and signaling pathways to maintain GA homeostasis in plants. In GA signal transduction, DELLA plays an important role as a negative regulator (Davière et al. 2008; Ueguchi-Tanaka et al. 2007). DELLA belongs to a subfamily of plant-specific GRAS proteins and is part of a highly conserved gene family. The main pathway of GA signaling depends on DELLA degradation (Davière et al. 2008). GID1 is a soluble gibberellin receptor that usually has a high binding affinity with biologically active GA but a low affinity with bioinert GA (Nakajima et al. 2006). GID1 combines with active GA to form the GID1-GA complex, while DELLA inhibits GA signaling and degrades upon interaction with the GID1-GA complex (Ueguchi-Tanaka et al. 2007). Additionally, the DELLA protein is involved in feedback regulation, regulating GA homeostasis by upregulating GA biosynthesis and gene expression of the GA receptor (Zentella et al. 2006). GID2 gene encodes a kind of F-box protein. As a positive regulator, it regulates GA signaling by controlling the stability of DELLA, a negative regulator (McGinnis et al. 2003). In this study, we found that DELLA was the core factor of the GA signaling pathway and participated in the feedback regulation of GA. The expression of GID1 and DELLA genes in dwarf plants was significantly higher than that in the control, inhibiting GA signaling and leading to the upregulation of GA3ox genes and downregulation of GA2ox genes. It was speculated that this may be the key process of dwarfing caused by GA signaling.
TF factor phytochrome-interacting factors (PILs/PIFs) play an essential role in the activation of elongation genes. DELLA could interact with the bHLH DNA-recognition domain and sequester these factors into an inactive complex to repress the transcriptional activity of PIF factors (De et al. 2008). However, in this study, TF was upregulated in dwarf banana. Light and hormones play an important role in cell expansion. In a growth regulatory program integrating light- and hormone-related pathways, PIFs act as the central hubs (De et al. 2014). Signaling by gibberellin (GA), brassinosteroid (BR) and auxin is integrated at the transcriptional level (Durbak et al. 2012). Therefore, it is necessary to combine the gene regulation of GA metabolism with that of BR and auxin for further discussion.
The Relationship Between IAA and Banana Dwarfism
Almost every aspect of plant life could be affected by auxin (Leyser 2018). The synthesis process of IAA is complex and involves the transformation of tryptophan (Trp) into IAA through a two-step catalytic reaction of the tryptophan aminotransferase of Arabidopsis (TAA) family of aminotransferases and the YUC family of flavin monooxygenases (YUCCA), which are part of the essential synthesis path of IAA (Zhao et al. 2012). YUCCA is the key gene in IAA metabolism (Kasahara 2016). In another synthesis pathway, tryptophan decarboxylase (TDC) family enzymes can convert Trp to tryptamine (TAM) (Mano and Nemoto 2012). The aldehyde oxidase (AAO) family catalyzes the conversion of indole-3-acetaldehyde (IAD) to IAA (Kasahara 2016; Mano and Nemoto 2012), and AAO is also the key gene in IAA metabolism. In this study, there were 9 differentially expressed genes related to IAA anabolism, 6 of which belong to the YUCCA gene family, but most of them were expressed at low levels. The two genes with relatively high expression did not have consistent change trends in the two experimental groups; both had higher expression in dwarf banana W1 but lower expression in dwarf banana W2. Moreover, DEGs related to TDC and AAO showed similar irregularities. Considering that the IAA contents in W1 and W2 were extremely significantly higher than those in H1 and H2, respectively (P < 0.01) (Fig. 1E), it was obvious that in the local biosynthesis of banana leaves, the differences in IAA contents were not caused by any of the abovementioned DEGs. The synthesis and metabolism of IAA may be regulated by other DEGs at the same time, thus affecting plant height.
Local auxin biosynthesis cooperates with the auxin transport mechanism to produce a strong morphogenetic auxin gradient to regulate plant growth and to interact with the changing environment. According to the classical view of auxin research, most auxin in plants is produced in the shoot tip meristem, young leaves and flower buds (Teale et al. 2006). Auxin is transported from the biosynthesis site to the action site through auxin efflux carriers, such as PIN-FORMED (PIN) protein (Kleine and Friml 2008). The AUX1/LAX family and ABCB subfamily are two other transporters of polar auxin (Cho and Cho 2013) that play an important role in the process of synthesis and transport coordination. In Arabidopsis, it was found that auxin transport between stems and roots could not compensate for local auxin deficiency in stems or roots, such as that due to the yucQ mutant (Chen et al. 2014). In this study, we found that there were 4 PIN genes and 8 ABCB genes with significant differences in expression (Table S1). Although it is not clear how auxin is transported in banana plants, it is obvious that the differences in transport mechanisms and in the expression of biosynthesis genes lead to different gradients of auxin, resulting in higher apoplastic auxin levels in dwarf banana and dynamic extracellular and intracellular IAA levels. However, the intracellular IAA concentration cannot be detected at present (Leyser 2018), so the mechanism cannot be confirmed. A similar phenomenon was observed in Arabidopsis thaliana; that is, the free IAA concentration in the dwarf mutant was not lower than that in the control (Li et al. 2009).
Aux/IAA encodes transcriptional repressors that respond to auxin changes and is a key component of auxin signaling (Mockaitis and Estelle 2008; Tian et al. 2018). Auxin functions by stimulating the ubiquitin-mediated proteolysis of Aux/IAAs (Teale et al. 2006). ARF proteins contain domains related to DNA binding, transcriptional activation or inhibition, and protein interaction (Tiwari et al. 2003). The degradation of Aux/IAA repressors depends on the level of auxin to affect the activity of ARF transcription factors (Chapman and Estelle 2009). Therefore, this process can activate the expression of target genes, which play an important role in auxin signal transduction. In this study, we found that most AUX/IAA genes were upregulated in dwarf plants, which inhibited the release of active ARF, leading to IAA deficiency and the dwarfing phenotype in plants. In addition, there were significant differences in the genes involved in IAA anabolism and signal transduction in both experimental groups, which may lead to differences in phenotype, such as the smaller leaf spacing in W2 than H2.
In conclusion, the IAA content in dwarf banana was significantly lower than that of the control material. The differences in IAA level and plant height were more likely caused by the differential expression of transporter proteins (such as PIN, ABCB and AUX1/LAX) and signal transducer genes (such as AUX/IAA). Differential expression of anabolic genes (such as YUCCA, DCC and AAO) may have a relatively weak impact.
The Relationship Between BRs and Banana Dwarfism
Facing the complicated impact caused by development and the environment, plants have different coping strategies; among them, BRs play fundamental roles. BRs regulate a series of biological processes, including photomorphogenesis, cell elongation, senescence, vasculature differentiation and stress responses (Yu et al. 2011). In general, cell expansion or elongation is the main growth-promoting role of BRs (Clouse and Sasse 1998).
In the two BR synthesis pathways, the key genes downstream were BR6ox1,2 and CYP92A6. Plants with loss-of-function mutations in these genes exhibit a dwarf phenotype (Kim et al. 2009; Shimada et al. 2003). In this study, we found that these genes showed irregular up- or downregulation, and there was no significant difference in BR hormone contents between dwarf banana and the control materials.
BRI1 and its homologous protein are receptor kinases of BR (He et al. 2005). Brassinolide (BL) is the first active BR to be isolated, and it can promote basal BRI1 kinase activity by binding to the extracellular domain of the BRI1 receptor kinase. The negative regulator BKI1, which acts on Y211, could be phosphorylated by BRI1 kinase. After being released from the membrane, it can associate with BAK1 or its homologs BKK1 and SERK1 (Clouse 2011). In this study, there was no obvious difference in the expression of genes lying downstream of BR signaling, which may be due to the interaction between BRI1 and BKI1. BRASSINAZOLE RESISTANT 1 (BZR1) and bri1-EMS-SUPPRESSOR 1 (BES1) are nuclear components of the BR signaling pathway. They mediate different kinds of responses in plants, including BR-induced growth responses and feedback regulation of BR biosynthesis (Wang et al. 2002). However, in this study, there was no significant difference between BZR1 and BES1. A similar finding was reported in which many target genes were not controlled by BZR1 and BES1 (Clouse 2011). The expression of TCH4 and CYCD3 in dwarf banana decreased significantly and may be affected by other factors.
The determination of hormone contents showed that the content of BRs in dwarf banana was higher than that in the corresponding control material (P > 0.05, Fig. 1D), so the dwarfism in this research is likely not caused by BR deficiency. On the other hand, the analysis of differential gene expression showed that the signal transduction of BR was not inhibited in dwarf banana, so the dwarfism in this research is likely not caused by insensitivity to BR. Therefore, BR may have little effect on banana dwarfism.
The Interactions Among the Three Hormones
Signaling pathways interconnect with each other in a complex network. This interconnection affects the growth and development of plants and is particularly important in hormone signaling (Blázquez et al. 2020), very often in response to the environment (Gallego et al. 2012). Hormone signaling cascades usually regulate their own biosynthesis, resulting in feedback regulation (Depuydt and Hardtke 2011).
Auxin can be used as a mobile signal to regulate the biosynthesis of GA (Frigerio and Alabadí 2006). Under the condition of low GA activity, auxin has been demonstrated to induce the expression of the GA biosynthesis gene GA20ox in tobacco and Arabidopsis, while in pea, auxin induces the expression of GA3ox and inhibits the expression of GA2ox (Weiss and Ori 2007). In this study, it was very likely that GA3ox and GA2ox were regulated by negative feedback under the combined influence of low concentrations of active GAs (GA1 and GA4) and extractive IAA, which was significantly higher in dwarf banana. Auxin could also activate the transcription of GA biosynthesis by degrading the transcriptional repressor of the GA signaling pathway, namely, DELLA (Salanenka et al. 2018). In turn, the interaction between the intermediate region of ARF6 and the DELLA protein repressor prevented the binding of ARF6 to the target DNA (Oh et al. 2014). In this study, ARF was significantly upregulated overall. However, DELLA was also upregulated at the same time, which may inhibit the interaction of ARF6 with its target DNA. Affected by the interaction between ARF6 and DELLA, CH4 and SAUR showed downregulation.
GA was also related to the transporter PIN. Insufficiency of GA can strongly inhibit the process of PIN transfer, while high levels of GA can promote PIN transporters (Löfke et al. 2013; Salanenka et al. 2018). However, the specific regulatory mechanism was not clear. In addition, ARF6 directly interacts with PIF4 through the bHLH domain, and they have many common target genes (Oh et al. 2014). Although it was not clear how they interact with each other, this interdependence provided a good model to explain the expression of ARF and PIF and the regulation of their target genes. In summary, in this study, IAA may participate in the feedback regulation of GA, while a low level of GA in turn inhibited the transport of IAA. The regulation between GA and IAA jointly regulated the hormone contents and the expression level of related genes, resulting in significant phenotypic differences between dwarf banana and the control.
BR signaling participates in the cell elongation promoted by GA; on the other hand, BR or active BZR1 could suppress the dwarf phenotype in GA-deficient plants. DELLA inhibited BZR1–DNA binding and directly interacted with BZR1 both in vitro and in vivo (Bai et al. 2012; Gallego et al. 2012). It has been shown that the BR and auxin signaling pathways interact with each other. BIN2 can phosphorylate the transcription factor AUXIN RESPONSE FACTOR-2 (ARF2) to inhibit its activity (Vert et al. 2008). In the two regulatory modules of BR interacting with GA and IAA, there was no difference in the BZR1 and BIN2 genes, which were the key genes in the BR signaling pathway. According to the results discussed above, BR had little effect on banana dwarfing. Moreover, the increase in auxin levels saturated the growth response stimulated by BR. As a result, the regulatory effect of BR on gene expression could be greatly reduced (Nemhauser et al. 2004). The increase in extracellular IAA may also reduce the effect of BR on gene expression. In conclusion, the anabolism and signal transduction of BR may not have a significant effect on the levels of GA and IAA or plant height.
Hypothesized Model of Banana Dwarfing
Based on the results of WGCNA, we further screened the DEGs related to hormone metabolism and signal transduction of GA and IAA and obtained a series of genes with a high correlation with hormone levels and plant height. We speculated that these genes may play a key role in dwarfing and hormone regulation, including GA2ox in the anabolic pathway of GA, DELLA and GID1 in the signal transduction pathway of GA, and TDC, YUCCA, AUX/IAA and SAUR in the anabolic pathway of IAA. Based on these key genes from WGCNA, we attempted to build a model to explain banana dwarfing (Fig. 11).
A schematic diagram of the hypothesized regulatory mechanism of banana dwarfing. Red represents upregulation, and green represents downregulation. PIN is shown in black because it does not represent all PIN transporters, and its regulatory mechanism is not clear. Solid lines represent the metabolic or signaling pathways, and dotted lines represent feedback regulation. The arrows represent positive regulation, and short lines represent negative regulation (Color figure online)
The differential expression of genes involved in GA anabolism led to a significant decrease in GA levels. Under the influence of anabolism and transporter DEGs, intracellular IAA was reduced significantly as a result of signal transduction. Then, the activities of PIF and ARF were inhibited by DELLA and AUX/IAA, respectively, which resulted in the inhibition of plant growth and led to banana dwarfing.
Conclusions
At present, the dwarfing mechanism of banana is not clear, and there is little correlational research. In this study, we investigated the relationship between hormones and banana dwarfism and revealed the metabolic regulatory mechanism and the differences in gene expression at the molecular level through transcriptome analysis.
According to the results, we inferred a complex regulatory network of banana dwarfing. First, in terms of endogenous hormone levels, GA (GA1, GA4) and IAA had significant effects on banana dwarfing, but BR had little effect. Second, in terms of anabolism and metabolic genes related to hormones, the genes most associated with banana plant height were GA2ox for GA and TDC and YUCCA for IAA. The differential expression of these genes was the main factor affecting hormone levels and plant height. Third, in terms of hormone signal transduction, DELLA and AUX/IAA repressor proteins were the core regulators of GA and IAA, respectively. They inhibited the process of signal transduction and had feedback regulation on hormone levels. It cannot be ignored that the transporter protein PIN, AUX1/LAX protein family and ABCB subfamily play an important role in the transport of IAA. In addition, it should be noted that the factors affecting plant height were complex and diverse. This study mainly explored the changes in hormone contents and gene expression in the young leaves of dwarf banana plants, which was completed without varying external factors (external application of hormones or inhibitors, light control, temperature control, etc.). Whole-plant analysis of plant hormones and the transcriptome was not the focus of this study. Further studies are worth pursuing to provide a reliable basis for the improvement of dwarf varieties.
References
Bai MY, Shang JX, Oh E et al (2012) Brassinosteroid, gibberellin and phytochrome impinge on a common transcription module in Arabidopsis. Nat Cell Biol 14(8):810–817. https://doi.org/10.1038/ncb2546
Blázquez MA, Nelson DC, Weijers D (2020) Evolution of plant hormone response pathways. Annu Rev Plant Biol. https://doi.org/10.1146/annurev-arplant-050718-100309
Chapman EJ, Estelle M (2009) Mechanism of auxin-regulated gene expression in plants. Annu Rev Genet 43:265–285. https://doi.org/10.1146/annurev-genet-102108-134148
Chen Q, Dai X, De-Paoli H et al (2014) Auxin overproduction in shoots cannot rescue auxin deficiencies in Arabidopsis roots. Plant Cell Physiol 55(6):1072–1079. https://doi.org/10.1093/pcp/pcu039
Chen J, Xie J, Duan Y et al (2016) Genome-wide identification and expression profiling reveal tissue-specific expression and differentially-regulated genes involved in gibberellin metabolism between Williams banana and its dwarf mutant. BMC Plant Biol 16(1):123. https://doi.org/10.1186/s12870-016-0809-1
Chiang HH, Hwang I, Goodman HM (1995) Isolation of the Arabidopsis GA4 locus. Plant Cell 7(2):195–201. https://doi.org/10.1105/tpc.7.2.195
Cho M, Cho HT (2013) The function of ABCB transporters in auxin transport. Plant Signal Behav 8(2):642–654. https://doi.org/10.4161/psb.22990
Clouse SD (2011) Brassinosteroid signal transduction: from receptor kinase activation to transcriptional networks regulating plant development. Plant Cell 23(4):1219–1230. https://doi.org/10.1105/tpc.111.084475
Clouse SD, Sasse JM (1998) Brassinosteroids: essential regulators of plant growth and development. Annu Rev Plant Biol 49(1):427–451. https://doi.org/10.1146/annurev.arplant.49.1.427
Damasco OP, Graham GC, Henry RJ et al (1996) Random amplified polymorphic DNA (RAPD) detection of dwarf off-types in micropropagated Cavendish (Musa spp. AAA) bananas. Plant Cell Rep 16(1–2):118–123. https://doi.org/10.1007/BF01275464
Dash PK, Rai R (2016) Translating the “Banana genome” to delineate stress resistance, dwarfing, parthenocarpy and mechanisms of fruit ripening. Front Plant Sci 7:1543. https://doi.org/10.3389/fpls.2016.01543
Davière JM, De Lucas M, Prat S (2008) Transcriptional factor interaction: a central step in DELLA function. Curr Opin Genet Dev 18(4):295–303. https://doi.org/10.1016/j.gde.2008.05.004
De Lucas M, Prat S (2014) PIF s get BR right: PHYTOCHROME INTERACTING FACTOR s as integrators of light and hormonal signals. New Phytol 202(4):1126–1141. https://doi.org/10.1111/nph.12725
De Lucas M, Daviere JM, Rodríguez-Falcón M et al (2008) A molecular framework for light and gibberellin control of cell elongation. Nature 451(7177):480–484. https://doi.org/10.1038/nature06520
Depuydt S, Hardtke CS (2011) Hormone signalling crosstalk in plant growth regulation. Curr Biol 21(9):R365–R373. https://doi.org/10.1016/j.cub.2011.03.013
Dill A, Sun T (2001) Synergistic derepression of gibberellin signaling by removing RGA and GAI function in Arabidopsis thaliana. Genetics 159(2):777–785
Durbak A, Yao H, McSteen P (2012) Hormone signaling in plant development. Curr Opin Plant Biol 15(1):92–96. https://doi.org/10.1016/j.pbi.2011.12.004
Frigerio M, Alabadí D, Pérez-Gómez J et al (2006) Transcriptional regulation of gibberellin metabolism genes by auxin signaling in Arabidopsis. Plant Physiol 142(2):553–563. https://doi.org/10.1104/pp.106.084871
Gallego-Bartolomé J, Minguet EG, Grau-Enguix F et al (2012) Molecular mechanism for the interaction between gibberellin and brassinosteroid signaling pathways in Arabidopsis. Proc Natl Acad Sci USA 109(33):13446–13451. https://doi.org/10.1073/pnas.111999210
Han F, Zhu B (2011) Evolutionary analysis of three gibberellin oxidase genesin rice, Arabidopsis, and soybean. Gene 473(1):23–35. https://doi.org/10.1016/j.gene.2010.10.010
He JX, Gendron JM, Sun Y et al (2005) BZR1 is a transcriptional repressor with dual roles in brassinosteroid homeostasis and growth responses. Science 307(5715):1634–1638. https://doi.org/10.1126/science.1107580
Hedden P (2003) The genes of the Green Revolution. Trends Genet 19(1):5–9. https://doi.org/10.1016/S0168-9525(02)00009-4
Hedden P, Kamiya Y (1997) Gibberellin biosynthesis: enzymes, genes and their regulation. Annu Rev Plant Biol 48(1):431–460. https://doi.org/10.1146/annurev.arplant.48.1.431
Kasahara H (2016) Current aspects of auxin biosynthesis in plants. Biosci Biotechnol Biochem 80(1):34–42. https://doi.org/10.1080/09168451.2015.1086259
Kim TW, Guan S, Sun Y et al (2009) Brassinosteroid signal transduction from cell-surface receptor kinases to nuclear transcription factors. Nat Cell Biol 11(10):1254–1260. https://doi.org/10.1016/j.phytochem.2004.01.007
Kleine-Vehn J, Friml J (2008) Polar targeting and endocytic recycling in auxin-dependent plant development. Annu Rev Cell Dev Biol 24:447–473. https://doi.org/10.1146/annurev.cellbio.24.110707.175254
Langfelder P, Horvath S (2008) WGCNA: an R package for weighted correlation network analysis. BMC Bioinformatics 9(1):559. https://doi.org/10.1186/1471-2105-9-559
Leyser O (2018) Auxin signaling. Plant Physiol 176(1):465–479. https://doi.org/10.1104/pp.17.00765
Li H, Cheng Y, Murphy A et al (2009) Constitutive repression and activation of auxin signaling in Arabidopsis. Plant Physiol 149(3):1277–1288. https://doi.org/10.1104/pp.108.129973
Löfke C, Zwiewka M, Heilmann I et al (2013) Asymmetric gibberellin signaling regulates vacuolar trafficking of PIN auxin transporters during root gravitropism. Proc Natl Acad Sci USA 110(9):3627–3632. https://doi.org/10.1073/pnas.1300107110
Ma Y, Xue H, Zhang L et al (2016) Involvement of auxin and brassinosteroid in dwarfism of autotetraploid apple (Malus× domestica). Sci Rep 6(1):1–14. https://doi.org/10.1038/srep26719
MacMillan J (2001) Occurrence of gibberellins in vascular plants, fungi, and bacteria. J Plant Growth Regul 20(4):387–442. https://doi.org/10.1007/s003440010038
Mano Y, Nemoto K (2012) The pathway of auxin biosynthesis in plants. J Exp Bot 63(8):2853–2872. https://doi.org/10.1093/jxb/ers091
Martin DN, Proebsting WM, Hedden P (1997) Mendel’s dwarfing gene: cDNAs from the Le alleles and function of the expressed proteins. Proc Natl Acad Sci 94(16):8907–8911. https://doi.org/10.1073/pnas.94.16.8907
McGinnis KM, Thomas SG, Soule JD et al (2003) The Arabidopsis SLEEPY1 gene encodes a putative F-box subunit of an SCF E3 ubiquitin ligase. Plant Cell 15(5):1120–1130. https://doi.org/10.1105/tpc.010827
Mockaitis K, Estelle M (2008) Auxin receptors and plant development: a new signaling paradigm. Annu Rev Cell Dev Biol. https://doi.org/10.1146/annurev.cellbio.23.090506.123214
Nakajima M, Shimada A, Takashi Y, Kim YC, Park SH et al (2006) Identification and characterization of Arabidopsis gibberellin receptors. Plant J 46:880–889. https://doi.org/10.1111/j.1365-313X.2006.02748.x
Nemhauser JL, Mockler TC, Chory J (2004) Interdependency of brassinosteroid and auxin signaling in Arabidopsis. PLoS Biol. https://doi.org/10.1371/journal.pbio.0020258
Nemhauser JL, Hong F, Chory J (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126(3):467–475. https://doi.org/10.1016/j.cell.2006.05.050
Oh E, Zhu JY, Bai MY et al (2014) Cell elongation is regulated through a central circuit of interacting transcription factors in the Arabidopsis hypocotyl. Elife 3:e03031. https://doi.org/10.7554/eLife.03031
Olszewski N, Sun T, Gubler F (2002) Gibberellin signaling: biosynthesis, catabolism, and response pathways. Plant Cell 14(suppl 1):S61–S80. https://doi.org/10.1105/tpc.010476
Phillips AL, Ward DA, Uknes S et al (1995) Isolation and expression of three gibberellin 20-oxidase cDNA clones from Arabidopsis. Plant Physiol 108(3):1049–1057. https://doi.org/10.1104/pp.108.3.1049
Richards DE, King KE, Ait-Ali T et al (2001) How gibberellin regulates plant growth and development: a molecular genetic analysis of gibberellin signaling. Annu Rev Plant Biol 52(1):67–88. https://doi.org/10.1146/annurev.arplant.52.1.67
Salanenka Y, Verstraeten I, Löfke C et al (2018) Gibberellin DELLA signaling targets the retromer complex to redirect protein trafficking to the plasma membrane. Proc Natl Acad Sci USA 115(14):3716–3721. https://doi.org/10.1073/pnas.1721760115
Shao X, Wu S, Dou T et al (2020) Using CRISPR/Cas9 genome editing system to create MaGA20ox2 gene-modified semi-dwarf banana. Plant Biotechnol J 18(1):17–19. https://doi.org/10.1111/pbi.13216
Shimada Y, Goda H, Nakamura A et al (2003) Organ-specific expression of brassinosteroid-biosynthetic genes and distribution of endogenous brassinosteroids in Arabidopsis. Plant Physiol 131(1):287–297. https://doi.org/10.1104/pp.013029
Sun T, Gubler F (2004) Molecular mechanism of gibberellin signaling in plants. Annu Rev Plant Biol 55:197–223. https://doi.org/10.1146/annurev.arplant.55.031903.141753
Teale WD, Paponov IA, Palme K (2006) Auxin in action: signalling, transport and the control of plant growth and development. Nat Rev Mol Cell Biol 7(11):847–859. https://doi.org/10.1038/nrm2020
Thomas SG, Phillips AL, Hedden P (1999) Molecular cloning and functional expression of gibberellin 2-oxidases, multifunctional enzymes involved in gibberellin deactivation. Proc Natl Acad Sci USA 96(8):4698–4703. https://doi.org/10.1073/pnas.96.8.4698
Tian H, Lv B, Ding T et al (2018) Auxin-BR interaction regulates plant growth and development. Front Plant Sci 8:2256. https://doi.org/10.3389/fpls.2017.02256
Tiwari SB, Hagen G, Guilfoyle T (2003) The roles of auxin response factor domains in auxin-responsive transcription. Plant Cell 15(2):533–543. https://doi.org/10.1105/tpc.008417
Ueguchi-Tanaka M, Nakajima M, Motoyuki A et al (2007) Gibberellin receptor and its role in gibberellin signaling in plants. Annu Rev Plant Biol 58:183–198. https://doi.org/10.1146/annurev.arplant.58.032806.103830
Vert G, Walcher CL, Chory J et al (2008) Integration of auxin and brassinosteroid pathways by Auxin Response Factor 2. Proc Natl Acad Sci 105(28):9829–9834. https://doi.org/10.1073/pnas.0803996105
Wang ZY, Nakano T, Gendron J et al (2002) Nuclear-localized BZR1 mediates brassinosteroid-induced growth and feedback suppression of brassinosteroid biosynthesis. Dev Cell 2(4):505–513. https://doi.org/10.1016/S1534-5807(02)00153-3
Wang B, Smith SM, Li J (2018) Genetic regulation of shoot architecture. Annu Rev Plant Biol 69:437–468. https://doi.org/10.1146/annurev-arplant-042817-040422
Wei C, Zhu L, Wen J et al (2018) Morphological, transcriptomics and biochemical characterization of new dwarf mutant of Brassica napus. Plant Sci 270:97–113. https://doi.org/10.1016/j.plantsci.2018.01.021
Weiss D, Ori N (2007) Mechanisms of cross talk between gibberellin and other hormones. Plant Physiol 144(3):1240–1246. https://doi.org/10.1104/pp.107.100370
Yamaguchi S (2008) Gibberellin metabolism and its regulation. Annu Rev Plant Biol 59:225–251. https://doi.org/10.1146/annurev.arplant.59.032607.092804
Yu X, Li L, Zola J et al (2011) A brassinosteroid transcriptional network revealed by genome-wide identification of BESI target genes in Arabidopsis thaliana. Plant J 65(4):634–646. https://doi.org/10.1111/j.1365-313X.2010.04449.x
Zentella R, Zhang ZL, Park M et al (2007) Global analysis of DELLA direct targets in early gibberellin signaling in Arabidopsis. Plant Cell 19(10):3037–3057. https://doi.org/10.1105/tpc.107.054999
Zhao Y (2012) Auxin biosynthesis: a simple two-step pathway converts tryptophan to indole-3-acetic acid in plants. Mol Plant 5(2):334–338. https://doi.org/10.1093/mp/ssr104
Acknowledgements
The authors are thankful to the Horticultural Research Institute, Guangxi Academy of Agricultural Sciences, Nanning, China, and the Nanning Investigation and Experiment Station of the Subtropical Fruits, Ministry of Agriculture, P. R. China, for research facilities.
Funding
This research was supported by the Science and Technology Major Project of Guangxi (Grant Numbers AA17204026, AA17204045) and the ‘948’ Project of Agricultural Ministry of China (Grant Numbers 2011-G16, 2016-X22).
Author information
Authors and Affiliations
Contributions
Conceptualization, Supervision, Writing—Review & Editing, J.Y. and B.D.; Methodology, Formal analysis, J.Y., B.D., X.L. and R.F.; Writing—Original Draft, Data Curation, Visualization, B.D. and X.W.; Software, Investigation, S.Z., J.Z., X.P. and Z.A.; Validation, Y.G., W.H. and W.T. All authors read and agreed with the final version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflicts of interest.
Additional information
Handling Editor: Theo Lange.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The following are available online at www.mdpi.com/xxx/s1, Fig. S1: Plant growth status at the first and fifth timepoints. Fig. S2: Comparison of RNA-seq data and genes in the reference genome. Fig. S3: GS value between each character and module in WGCNA. Fig. S4: Venn diagram of core genes in WGCNA. Table S1: The differential expression and annotation of all DEGs in this study. Table S2: Sequences of primers used in qRT-PCR.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Deng, B., Wang, X., Long, X. et al. Plant Hormone Metabolome and Transcriptome Analysis of Dwarf and Wild-type Banana. J Plant Growth Regul 41, 2386–2405 (2022). https://doi.org/10.1007/s00344-021-10447-7
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00344-021-10447-7