Abstract
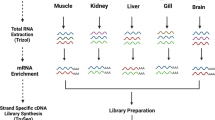
At present, the function and profile of long noncoding RNAs (IncRNAs) in half-smooth tongue soles Cynoglossus semilaevis remain poorly understood. Lherefore, we identified IncRNAs in the species using large-scale deep sequencing approaches. A total of 96 726 222 clean reads and 2497 IncRNAs were obtained from the compound samples. In total, 1 694 known IncRNAs and 803 novel IncRNAs were identified. Most of the novel IncRNAs distributed mainly in a range of 200–3 000 nt and contained 2–6 exons. Six novel IncRNAs were selected for expression analysis by quantitative reverse transcription-polymerase chain reaction, of which lnc_770 and lnc_150 were expressed mainly in the ovary. Lhis study provides a valuable resource for IncRNA studies of half-smooth tongue sole and improves our understanding of the function of non-coding RNA in fish.
Similar content being viewed by others
References
Chen S L, Tian Y S, Yang J F, Shao C W, Ji X S, Zhai J M, Liao X L, Zhuang Z M, Su P Z, Xu J Y Sha Z X, Wu P F, Wang N. 2009. Artificial gynogenesis and sex determination in half-smooth tongue sole (Cynoglossus semilaevis). Marine Biotechnology), 11(2): 243–251.
Chen S, Zhang G J, Shao C W, Huang Q F, Liu G, Zhang P Song W T, An N, Chalopin D, Volff J N, Hong Y H, Li Q Y Sha Z X, Zhou H L, Xie M S, Yu Q L, Liu Y, Xiang H, Wang N, Wu K, Yang C G, Zhou Q, Liao X L, Yang L F Hu Q M, Zhang J L, Meng L, Jin L J, Tian Y S, Lian J M, Yang J F, Miao G D, Liu S S, Liang Z, Yan F, Li Y Z, Sun B, Zhang H, Zhang J, Zhu Y, Du M, Zhao Y W, Schartl M, Tang Q S, Wang J. 2014. Whole-genome sequence of a flatfish provides insights into ZW sex chromosome evolution and adaptation to a benthic lifestyle. Nature Genetics, 46(3): 253–260.
Cui Z K, Liu Y Wang W W, Wang Q, Zhang N, Lin F, Wang N, Shao C W Dong Z D, Li Y Z, Yang Y M, Hu M Z, Li H L, Gao F T, Wei Z F, Meng L, Liu Y, Wei M, Zhu Y, Guo H, Cheng CHK, Schartl M, Chen S L. 2017. Genome editing reveals dmrt1 as an essential male sex-determining gene in Chinese tongue sole (Cynoglossus semilaevis) Scientific Reports, 7: 42 213.
Edgar R, Domrachev M, Lash A E. 2002. Gene expression omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Research, 30(1): 207–210.
Gong C G, Li Z Z, Ramanujan K, Clay I, Zhang Y Y, Lemire-Brachat S, Glass D J. 2015. A long non-coding RNA, LncMyoD, regulates skeletal muscle differentiation by blocking IMP2-mediated mRNA translation. Development Cell, 34 (2): 181–191.
Jiang X Y Ning Q L. 2015. The emerging roles of long noncoding RNAs in common cardiovascular diseases. Hypertension Research, 38(6): 375–379.
Kong L, Zhang Y Ye Z Q, Liu X Q, Zhao S Q, Wei L P, Gao G. 2007. CPC: assess the protein-coding potential of transcripts using sequence features and support vector machine. Nucleic Acids Research, 35(S2): W345–W349.
Li J H, Lv Y Liu R R, Yu Y Shan C M, Bian W H, Jiang J, Zhang D L, Yang C, Sun Y Y. 2018. Identification and characterization of a conservative W chromosome-linked circRNA in half-smooth tongue sole (Cynoglossus semilaevis) reveal its female-biased expression in immune organs. Fish & Shellfish Immunology, 82: 1–531.
Liu J X, Zhang W, Du X X, Jiang J J, Wang C L, Wang X B, Zhang Q Q, He Y. 2016a. Molecular characterization and functional analysis of the GATA4 in tongue sole (Cynoglossus semilaevis). Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 193: 1–1.
Liu J X, Zhang W, Sun Y, Wang Z G, Zhang Q Q, Wang X B. 2016b. Molecular characterization and expression profiles of GATA6 in tongue sole (Cynoglossus semilaevis). Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 198: 1–19.
Lorenzen J M, Thum T. 2016. Long noncoding RNAs in kidney and cardiovascular diseases. Nature Reviews Nephrology, 12(6): 360–373.
Lu X K, Chen X G, Mu M, Wang J J, Wang X G, Wang D L, Yin Z J, Fan W L, Wang S, Guo L X, Ye W W. 2016. Genome-wide analysis of long noncoding RNAs and their responses to drought stress in cotton (Gossypium hirsutum L.). PLoS One, 11(6): e0156723.
Ma Y L, Yang Y Z, Wang F, Moyer M P, Wei Q, Zhang P, Yang Z, Liu W J, Zhang H Z, Chen N W, Wang H, Wang H M, Qin H L. 2016. Long non-coding RNA CCAL regulates colorectal cancer progression by activating Wnt/p-catenin signalling pathway via suppression of activator protein 2a. Gut, 65(9): 1494–1504.
Philippen L E, Dirkx E, da Costa-Martins P A, De Windt L J. 2015. Non-coding RNA in control of gene regulatory programs in cardiac development and disease. Journal of Molecular and Cellular Cardiology, 89: 1–51.
Roberts T C, Morris K V, Wood M J A. 2014. The role of long non-coding RNAs in neurodevelopment, brain function and neurological disease. Philosophical Transactions of the Royal Society B: Biological Science, 369(1652): pii: 20130507.
Santosh B, Varshney A, Yadava P K. 2015. Non-coding RNAs: biological functions and applications. Cell Biochemistry & Function, 33(1): 14–22.
Sauvageau M, Goff L A, Lodato S, Bonev B, Groff A F, Gerhardinger C, Sanchez-Gomez D B, Hacisuleyman E, Li E, Spence M, Liapis S C, Mallard W, Morse M, Swerdel M R, DEcclessis M F, Moore J C, Lai V, Gong G C, Yancopoulos G D, Frendewey D, Kellis M, Hart R P, Valenzuela D M, Arlotta P, Rinn J L. 2013. Multiple knockout mouse models reveal lincRNAs are required for life and brain development, eLife, 2: e01749.
Shao C W, Li Q Y, Chen S L, Zhang P, Lian J M, Hu Q M, Sun B, Jin L J, Liu S S, Wang Z J, Zhao H M, Jin Z H, Liang Z, Li Y Z, Zheng Q M, Zhang Y, Wang J, Zhang G J. 2014. Epigenetic modification and inheritance in sexual reversal offish. Genome Research, 24(4): 604–615.
Sun L, Luo H T, Bu D C, Zhao G G, Yu K T, Zhang C H, Liu Y N, Chen R S, Zhao Y 2013. Utilizing sequence intrinsic composition to classify protein-coding and long non-coding transcripts. Nucleic Acids Research, 41(17): e166.
Sun M, Kraus W L. 2015. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocrine Review, 36(1): 25–64.
Sun Y Y, Yu H Y, Zhang Q Q, Qi J, Zhong Q W, Chen Y J, Li C M. 2010. Molecular characterization and expression pattern of two zona pellucida genes in half-smooth tongue sole (Cynoglossus semilaevis). Comparative Biochemistry and Physiology Part B: Biochemistry and Molecular Biology, 155(3): 316–321.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley D R, Pimentel H, Salzberg S L, Rinn J L, Pachter L. 2012. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nature Protocols, 7(3): 562–578.
Viereck J, Kumarswamy R, Thum T. 2015. Long noncoding RNAs as inducers and terminators of vascular development. Circulation, 131(14): 1236–1238.
Wang L G, Park H J, Dasari S, Wang S Q, Kocher J P, Li W. 2013. CPAT: coding-potential assessment tool using an alignment-free logistic regression model. Nucleic Acids Research, 41(6): e74.
Xu W T, Li H L, Dong Z D, Cui Z K, Zhang N, Meng L, Zhu Y, Liu Y Li Y Z, Guo H, Ma J L, Wei Z F, Zhang N W, Yang Y M, Chen S L. 2016. Ubiquitin ligase gene neurl3 plays a role in spermatogenesis of half-smooth tongue sole (Cynoglossus semilaevis) by regulating testis protein ubiquitination. Gene, 592(1): 215–220.
Yan B, Yao J, Liu J Y, Li X M, Wang X Q, Li Y J, Tao Z F, Song Y C, Chen Q, Jiang Q. 2015. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circulation Research, 116(7): 1143–1156.
Yan H, Chen Y D, Zhou S, Li C, Gong G Y, Chen X J, Wang T Z, Chen S L, Sha Z X. 2016. Expression Profile Analysis of miR-221 and miR-222 in Different Tissues and Head Kidney Cells of Cynoglossus semilaevis, Following Pathogen Infection. Marine Biotechnology, 18(1): 37–48.
Yue F, Cheng Y, Breschi A, Vierstra J, Wu W S, Ryba T, Sandstrom R, Ma Z H, Davis C, Pope B D, Shen Y Pervouchine D D, Djebali S, Thurman R E, Kaul R, Rynes E, Kirilusha A, Marinov G K, Williams B A, Trout D, Amrhein H, Fisher-Aylor K, Antoshechkin I, DeSalvo G, See L H, Fastuca M, Drenkow J, Zaleski C, Dobin A, Prieto P, Lagarde J, Bussotti G, Tanzer A, Denas O, Li K W, Bender M A, Zhang M H, Byron R, Groudine M T, McCleary D, Pham L, Ye Z, Kuan S, Edsall L, Wu Y C, Rasmussen M D, Bansal M S, Kellis M, Keller C A, Morrissey C S, Mishra T, Jain D, Dogan N, Harris R S, Cayting P, Kawli T, Boyle A P, Euskirchen G, Kundaje A, Lin S, Lin Y, Jansen C, Malladi V S, Cline M S, Erickson D T, Kirkup V M, Learned K, Sloan C A, Rosenbloom K R, De Sousa B L, Beal K, Pignatelli M, Flicek P, Lian J, Kahveci T, Lee D, Kent W J, Santos M R, Herrero J, Notredame C, Johnson A, Vong S, Lee K, Bates D, Neri F, Diegel M, Canfield T, Sabo P J, Wilken M S, Reh T A, Giste E, Shafer A, Kutyavin T, Haugen E, Dunn D, Reynolds A P, Neph S, Humbert R, Hansen R S, De Bruijn M, Selleri L, Rudensky A, Josefowicz S, Samstein R, Eichler E E, Orkin S H, Levasseur D, Papayannopoulou T, Chang K H, Skoultchi A, Gosh S, Disteche C, Treuting P, Wang Y L, Weiss M J, Blobel G A, Cao X Y, Zhong S, Wang T, Good P J, Lowdon R F, Adams L B, Zhou X Q, Pazin M J, Feingold E A, Wold B, Taylor J, Mortazavi A, Weissman S M, Stamatoyannopoulos J A, Snyder M P, Guigo R, Gingeras T R, Gilbert D M, Hardison R C, Beer M A, Ren B, The Mouse ENCODE Consortium. 2014. A comparative encyclopedia of DNA elements in the mouse genome. Nature, 515(7527): 355–364.
Author information
Authors and Affiliations
Corresponding author
Additional information
Supported by the Shandong Provincial Natural Science Foundation (No. ZR2013CL003), the National Natural Science Foundation of China (Nos. 81670398, 91639102), and the funding of Taishan Scholars of Shandong Province to Binzhou Medical University and the Key Research Project of Yantai City (No. 2018ZHGY071)
Rights and permissions
About this article
Cite this article
Jiang, X., Jing, X., Lü, Y. et al. Genome-wide identification and prediction of long non-coding RNAs in half-smooth tongue sole Cynoglossus semilaevis. J. Ocean. Limnol. 38, 226–235 (2020). https://doi.org/10.1007/s00343-019-8287-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00343-019-8287-6