Abstract
Countries in the Western Indian Ocean (WIO) and along the Red Sea are particularly vulnerable to coral reef degradation, and understanding the degree of connectivity among coral reefs is a first step toward efficient conservation. The aim of this study is to investigate the genetic diversity, population structure and connectivity patterns of the broadcast spawning coral Acropora tenuis, first at a large scale comparing the Red Sea and the WIO, and second at a smaller scale comparing sites within the WIO. In total 689 individual A. tenuis colonies were sampled on 28 locations in Saudi Arabia, Kenya, Tanzania, Mozambique and Madagascar and analysed with seven microsatellite markers. The sample site in the Red Sea was found to be differentiated from all other sites in the WIO, which confirms the hypothesised genetic break. High differentiation was found between the African mainland and Madagascar and within Madagascar. However, there is evidence for long-distance larval dispersal for A. tenuis in the North Mozambique Channel region, with exchange between northern Mozambique and northern Madagascar. The sites in the southwest of Madagascar show mixing with sites in northern Madagascar, as well as exchange with sites in northern Mozambique and Tanzania. Southern Mozambique forms a separate group in Bayesian clustering. High genetic connectivity was found for most sites along the East African mainland coast, with no indication for strict genetic barriers. These results support biophysical modelling studies, which propose Tanzania as a seeding source of larvae for downstream Kenya. These patterns of high genetic connectivity combined with contemporary dispersal barriers can be explained by the long larval duration of A. tenuis and the prevailing northbound East African Coast Current that facilitates higher genetic connectivity along the northern East African Coast, while eddies in the Mozambique Channel are causing larval retention in southern Mozambique and Madagascar.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Understanding the degree of connectivity among coral reefs is a first step toward efficient conservation (Almany et al. 2009), as connectivity between reefs determines gene flow, genetic diversity and genetic structure of populations, as well as the capacity to persist under predicted climate change (Palumbi 2003). Coral connectivity, or the ‘movement of individuals within and among local or subpopulations’ (Cowen and Sponaugle 2009), is facilitated by dispersal of coral larvae. The Pelagic Larval Duration (PLD, i.e. the maximum time the larvae spend in the water column) and larval behaviour, but also environmental conditions, such as the suitability of the habitat for larval settling, currents and oceanographic barriers, determine the connectivity of coral reefs (Graham et al. 2008; Cowen and Sponaugle 2009). In theory, pelagic larval duration is positively correlated with dispersal distance, and longer PLD increases the number of connected coral reef sites in the Western Indian Ocean (WIO) (Crochelet et al. 2016), but see Weersing and Toonen (2009).
Increased knowledge on connectivity is becoming crucial as coral reefs are degrading at an alarming rate, due to a variety of local and global anthropogenic stressors (Hoegh-Guldberg et al. 2017; Hughes et al. 2017), thereby threatening an essential habitat of at least a quarter of known marine species (Fisher et al. 2015). Coral reefs also sustain an estimated value of around $352,000 ha−1 per year of ecological goods and services, such as food, coastal protection and recreation for over 500 million people (Moberg and Folke 1999; Costanza et al. 2014). On the one hand, developing countries in the WIO and countries along the Red Sea are particularly vulnerable to coral reef degradation (e.g. overfishing and destructive fishing, pollution and coastal development), with very low economic adaptive capacity to recover from coral reef loss (Burke et al. 2011). On the other hand, coral reefs in the WIO are some of the least studied in the world. Marine Ecoregions of the World by Spalding et al. (2007), and Ecoregions as proposed by Obura (2012), describe two scenarios of regional biogeographic patterns. Within-species connectivity data form an essential tool to verify and explain biogeographic regions described for corals in the WIO and Red Sea. Variable but consistently high connectivity was found on a small spatial scale in Kenya and Tanzania for A. tenuis (van der Ven et al. 2016), but information on connectivity on a larger spatial scale within the WIO (Fig. 1a), and between the WIO and the Red Sea (Fig. 2), is lacking. Acropora tenuis (Dana (1846); Cnidaria; Scleractinia; Acroporidae) is a branching scleractinian coral, which grows on upper reef slopes and has a widespread distribution in the Indo-Pacific, commonly occurring in the Red Sea, Madagascar, and along the coast of East Africa in Kenya and Tanzania, as well as in the north and south of Mozambique (Veron 2000). This species is a broadcast spawning coral which releases buoyant eggs and sperm to the water column where fertilisation takes place. Information on the spawning of acroporids in East Africa is limited (Baird et al. 2021), but seems to take place in and around the time of the Northeast Monsoon (currents depicted in Fig. 1). Indications for synchronised mass-spawning between September and December was found in Vamizi (Mozambique), Andavadoaka (Madagascar) and in Reunion (Sola et al. 2016; Baird et al. 2021), while unsynchronised spawning over a time span of seven months (from October to April) was observed on some occasions in Mombasa, Kenya (Mangubhai and Harrison 2008, 2009). In Saudi Arabia in the Red Sea acroporids were observed to spawn between April and June (Bouwmeester et al. 2015; Baird et al. 2021).
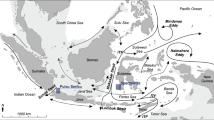
Map of the Western Indian Ocean. Major ocean currents during the Northeast Monsoon are indicated schematically (Schott and McCreary 2001; Sofianos and Johns 2003; Lutjeharms and Bornman 2010; Hancke et al. 2014). SEC: South Equatorial Current; NEMC: Northeast Madagascar Current; SEMC: Southeast Madagascar Current; EACC: East African Coast Current; SECC: South Equatorial Counter Current; SC: Somali Current; AC: Agulhas Current. Map showing a Marine Ecoregions of the world (Spalding et al. 2007): 95: East African Coral Coast; 99: Southeast Madagascar; 100: Western and Northern Madagascar; 101: Sofala; 102: Delagoa. b Ecoregions as suggested by Obura (2012): EAC: East African Coast region; NMC: Northern Mozambique Channel region; SMC: Southern Mozambique Channel region; EM: Eastern Madagascar; SM: Southern Mozambique
Map showing a sample sites of Acropora tenuis in Saudi Arabia, Kenya, Tanzania, Mozambique and Madagascar. Major ocean currents during the Northeast Monsoon are indicated schematically (Schott and McCreary 2001; Sofianos and Johns 2003; Lutjeharms and Bornman 2010; Hancke et al. 2014): SEC: South Equatorial Current; NEMC: Northeast Madagascar Current; SEMC: Southeast Madagascar Current; EACC: East African Coast Current; SECC: The South Equatorial Counter Current; SC: Somali Current; AC: Agulhas Current; sample site codes see Table 1. b Pie charts on the map show the distribution of five genetic clusters (K = 5) at sample sites as revealed in Bayesian clustering implemented in STRUCTURE (Pritchard et al. 2000) for sample sites in the Western Indian Ocean (WIO) only. c K = 8 bar plot for individuals from the Red Sea and WIO. The colours in the bar plot visualise the individual’s estimated membership fraction in each of the K clusters inferred by Bayesian clustering with the programme STRUCTURE, with each vertical bar representing one individual
The PLD, which potentially affects the capacity for long-distance dispersal, is highly variable in acroporids (e.g. between 54 and 209 days, Nishikawa and Sakai 2005; Graham et al. 2008). Acropora tenuis larvae are viable and can settle up to 69 days after spawning in laboratory experiments and have therefore the potential to disperse over long distances (Nishikawa et al. 2003). However, for all acroporids a high early mortality is found and most settlement occurs on the natal reef or adjacent reefs (Ayre and Hughes 2000; Graham et al. 2008; Underwood et al. 2009). Besides sexual reproduction through gametes, acroporid corals can also reproduce asexually (clonal reproduction) by colony fragmentation.
The aim of this study is to investigate the genetic diversity, population structure and connectivity patterns of the broadcast spawning coral A. tenuis first at a large scale, comparing the Red Sea and the WIO, and second at a smaller scale, comparing sites within the WIO. Based on multispecies distribution records of corals, six regions can be distinguished in the central WIO (Obura 2012; Fig. 1b): (1) the reefs surrounding the Northern Mozambique Channel (NMC region) that form the high-diversity core region in the Western Indian Ocean, (2) the East African Coast (EAC) region under influence of the East African Coast Current with diversity decreasing with increasing latitude (McClanahan et al. 2014), (3) the Southern Mozambique Channel region (SMC) under influence of Mozambique Channel eddies, (4) the Delagoa region under influence of the Agulhas Current, (5) East Madagascar (EM) under influence of the Northeast and Southeast Madagascar Current and (6) the Southern Mozambique region (SM). Four hypotheses are proposed based on previous biogeographic regions suggested in Obura (2012), ocean currents and the life history of A. tenuis: (I) a genetic break between the Red Sea and the WIO, (II) a distinct NMC region with higher genetic diversity, (III) a distinct SMC region, (IV) distinct regions of EM and SM in Madagascar. The hypotheses are tested with seven microsatellite markers.
Materials and methods
Coral reefs
Fringing reefs line the coast of the WIO, with the most extensive reefs found in the south of Tanzania and the north of Mozambique, and smaller and more fragmented reefs found in the north of Kenya and south of Mozambique (Obura 2012; McClanahan et al. 2014; Obura et al. 2017). The central Mozambique coastline is characterised by large-scale river outlets and mangrove forests and no coral reefs occur here, because of freshwater and sediment discharge (Obura et al. 2017). The Red Sea is a peripheral region of the Indo-Pacific and connected with the Gulf of Aden in the Indian Ocean through the shallow (130 m deep) and narrow strait of Bab-el-Mandab. The Red Sea has mainly fringing reefs along most of its shores, with atolls and pinnacle reefs found in the Southern and Central Red Sea (Wilkinson 2002).
Sampling
A total of 689 individual A. tenuis colonies were sampled (Fig. 2a, Table 1 and Appendix 1) in one location in Saudi Arabia, six locations in Mozambique and ten locations in Madagascar, while samples from five locations in Kenya and six locations in Tanzania were included from van der Ven et al. (2016). Colonies were randomly selected while maintaining at least two metres between them to avoid sampling clones. Each sample was obtained by breaking a 2–3 cm fragment from a branch. The fragments were stored in a plastic container with seawater until fixation in absolute ethanol. Samples were stored in the dark at 7 °C until extraction.
DNA extraction
The fragments, both skeleton and tissue together, were crushed manually with a bone cutter and incubated overnight at 56 °C with lysis buffer and Proteinase K. The NucleoSpin® Tissue DNA extraction kit (Macherey–Nagel, Düren, Germany) was used following the company’s animal tissue protocol for 100 µL DNA extract. The purity and concentration of the DNA extract was determined with a NanoDrop 2000 spectrophotometer (Thermo-scientific).
Microsatellite genotyping
Samples were genotyped using seven microsatellite primer pairs designed for Acropora millepora (Appendix 2), but cross-amplifications showed that they can be applied to A. tenuis (Underwood et al. 2009; van der Ven et al. 2016) and several other acroporid species from the Pacific Ocean (Richards and van Oppen 2012). The primers were combined in one multiplex (PCR) with a volume of 12.5 µL containing 2.5 µL template DNA, 1.25 µL (2 µM) primer mix with fluorescently labelled primers 6-FAM, VIC, PET and NED (Applied Biosystems, Foster City, CA, USA), 2.5 µL H2O and 6.25 µL QIAGEN Multiplex PCR Master Mix (QIAGEN, Hilden, Germany). The PCR cycling parameters were: an activation step for 5 min at 95 °C, followed by 35 cycles of denaturation for 30 s at 95 °C, annealing for 90 s at 60 °C and 30 s at 72 °C. The final extension was done for 30 min at 68 °C. The PCR product was diluted 10 or 100 times and analysed together with a GeneScantm-500 LIZ® size standard (Applied Biosystems, Foster City, CA, USA) on an ABI 3730 DNA Analyzer (Applied Biosystems, Foster City, CA, USA). The resulting electropherograms were scored manually with GeneMarker® (v. 2.4.0; SoftGenetics, State College, PA, USA).
Genetic and genotypic diversity analysis
The software GenAlEx 6.5 (Peakall and Smouse 2012) was used to identify multilocus genotypes (MLGs) that were likely to be the result of fragmentation of the coral colony. Clonal richness (R) was calculated as R = (G-1)/(N-1) in which N is the total number of samples, and G is the number of MLGs (Dorken and Eckert 2001). The departure from Hardy–Weinberg equilibrium (HWE) was determined for each locus with FSTAT 2.9.3 (Goudet 1995) and presented as FIS (calculated as smallF; Weir and Cockerham 1984), with significance levels based on 210,000 permutations. In all FSTAT analyses, the number of permutations was automatically determined based on the number of loci and populations in the experimental design. Populations with heterozygote deficiency were further analysed with INEst 2.0 (Chybicki and Burczyk 2009) utilising a Bayesian approach for estimating both null alleles and inbreeding simultaneously (Campagne et al. 2012). The model was run with 50,000 burn-ins and 500,000 cycles. Linkage disequilibrium (LD) was calculated with FSTAT, and the P value for genotypic disequilibrium was based on 6,300,000 permutations. Number of alleles per population, corrected for sample size and expressed as allelic richness, was also calculated with FSTAT. Private alleles and observed, expected and unbiased (sample size corrected) heterozygosity were calculated with GenAlEx.
Population structure
To investigate population structure among populations, pairwise FST values were estimated using the method of Weir and Cockerham (1984) (θ) with FSTAT and tested for significance with 55,000 permutations. In GenAlEx DEST (Jost 2008) was calculated and tested for significance with 9999 permutations. Pairwise FST values as well as DEST values were utilised in a Mantel test in GenAlEx to test for linear association with geographic distances measured as the shortest distance by sea (isolation by distance: IBD) with 9,999 permutations. The pairwise geographic distance was measured as the shortest path possible by water between two sample sites in Google Earth Pro (www.google.com/earth). An analysis of molecular variance (AMOVA) was conducted with GenAlEx, estimating overall FST as well as testing two potential population structures: Marine Ecoregions of the world as described by Spalding et al. (2007), and Ecoregions as proposed by Obura (2012), Fig. 1a and 1b, respectively. Significance was tested with 9,999 permutations. SPAGEDI (Hardy and Vekemans 2002) was used to test if the assumption of step-wise mutations was met (in this case RST would reflect population differentiation better), as well as to investigate the relative importance of mutation rates versus migration rates on population differentiation in distance intervals, tested with 20,000 permutations. Principal coordinates analyses (PCoA) were conducted in GenAlEx to examine the spatial variation based on pairwise FST. The software STRUCTURE 2.3.4 (Pritchard et al. 2000), which uses a Bayesian clustering based on the Markov chain Monte Carlo (MCMC) assignment method, was run under the admixture model to determine the number of genetic clusters (K) and was run both with and without prior population information. STRUCTURE analysis was performed in the ParallelStructure package (Besnier and Glover 2013) in R (https://www.r-project.org/). The STRUCTURE analysis was run for all data K = 1–28, as well as for the Western Indian Ocean excluding the Red Sea (K = 1–27). Analyses included 10 runs with a burn-in length of 100,000 and 1,000,000 MCMC replications. All analyses were implemented in the CIPRES Science Gateway portal (Miller et al. 2015). HARVESTER 0.6.94 (Earl and Vonholdt 2012) was used to visualise and analyse the STRUCTURE output by plotting log probability L (K) and ΔK (Evanno et al. 2005).
Additionally, all STRUCTURE analyses were repeated in InStruct to verify if the population structure found in STRUCTURE is affected by inbred populations (Gao et al. 2007). Lastly, a one-sided comparison (9,999 permutations) of HO, Ar, FIS and FST was made (FSTAT) to test whether these parameters were significantly different between the regions in the WIO.
Results
Genotypic diversity
Of the 689 sampled individuals, 629 were found to be unique multilocus genotypes (MLGs). Clonal richness (R) was mostly found to be the maximum value of 1 (Table 1), which indicates absence of clonality, with all different samples analysed corresponding to distinct lineages. Lower values of R were found in the south of Madagascar in Fa, Lv and FD (0.3, 0.3 and 0.5, respectively). As the probability of identity (PI) was low (< 0.001 for all sample sites, below the value of 0.01 set for adequate population studies (Waits et al. 2001)), the chance of colonies accidentally sharing the same genotype is very low. Therefore, the colonies sharing the same genotype were considered as potentially belonging to the same individual. Further analysis was performed with a reduced dataset, with all but one of the duplicated individuals from every identical MLG, in total 60 samples, excluded.
Genetic diversity
The seven microsatellite loci amplified successfully and were polymorphic for all 28 sample sites with 2–12 alleles per locus (Appendix 3). Allelic richness (Ar) ranged between 1.5 and 1.8, observed heterozygosity (HO) between 0.212 and 0.452 and unbiased expected heterozygosity (uHE) between 0.467 and 0.788 (Table 1). Private alleles were found along the whole range with a maximum of four in Ja and in low frequency with the highest in Nt (36% of individuals carried a private allele on locus Amil5_028). Separate analysis of the Red Sea, Madagascar and other sample sites revealed 1, 8 and 19 private alleles, respectively. Significant linkage disequilibrium (LD) was only found for two of the 21 locus pairs (P < 0.05) in sites La and Mk (data not shown). All sample sites showed positive FIS values ranging between 0.320 and 0.634 indicating heterozygotic deficits, and significant departure from Hardy–Weinberg equilibrium (HWE) (Table 1). Heterozygote deficits were observed for all loci in the WIO, and significant for all seven loci within the samples from sites Wa and Vi, and for six loci from RM, La, Mo, Di, Ki, Ja, Na, Ba, and Nb (Appendix 3). Further analysis with INEst supported a model including inbreeding for almost all sample sites, likely causing the high inbreeding coefficients. However, null alleles may be present in Ra and Nt (Appendix 4).
Population structure
Significant overall differentiation was found (FST = 0.128, P < 0.001), as well as in 53% of the pairwise comparisons of FST and 94% of pairwise comparisons of DEST (Table 2), with sites with small sample sizes (Mf, Pe, To, An, Lv) displaying more non-significant comparisons. Moderate but significant genetic differentiation (pairwise FST values) was found for the Red Sea site RM (0.018–0.313) versus all other sites. Within the WIO, high differentiation was found for sites in Madagascar, especially for site Ra (0.110–0.381), To (0.131–0.416) and Lv (0.176–0.461) versus all sites. Among the sites from the East African mainland the differentiation was lower, with the highest differentiation found for sites in Mozambique, for instance Va (0.088–0.316) and Vi (0.066–0.318). Although the pattern of differentiation is congruent with the results based on FST, DEST was consistently higher than the FST values, especially for pairwise comparisons with sites in Madagascar, while relatively lower values of DEST were found for the East African mainland sites.
The pairwise FST and DEST did not vary linearly with the geographic distance, and the mantel test showed no significant correlation between population differentiation and geographic distance for all sites in the Red Sea and Western Indian Ocean, Madagascar or the sites along mainland East Africa. However, there was isolation by distance when tested within the WIO (IBD FST; R2 = 0.0583, P < 0.05; IBD DEST; R2 = 0.088, P < 0.05; Appendix 5). No significant contribution of step-wise mutations (RST > FST) was found when testing the Red Sea and the WIO (interval 1–4000 km and interval 4001–6900 km), but significant contribution of step-wise mutations was found for distances beyond 1300 km within the WIO (Appendix 6).
Bayesian clustering in STRUCTURE which included a priori population information of the six ecoregions defined in the WIO (Fig. 1b) plus the Red Sea, resulted in a cluster distribution congruent with some of the predefined groups (especially the NMC and SMC, Appendix 7). However, this approach does come with the possibility that real population differences (within the defined groups) may be overlooked (Pritchard et al. 2000). Indeed, analyses without a priori population information revealed additional and alternative genetic structuring. For these latter analyses, a plateau for the log probability for the values of K ranging from 7 to 9 was found, of which K = 8 was selected (Appendix 8 and Fig. 2c), but the additional K’s were also included in the investigation. Within the WIO clustering revealed a plateau for the values of K ranging from 5 to 9, of which K = 5 was selected (Appendix 8, Fig. 2b). All clusters occurred at all sample sites, but based on the occurrence and proportion of the different genetic clusters within each sample site, groups of sample sites could be identified. A separate group can be distinguished in Saudi Arabia (RM), although some of the individuals might belong to different genetic clusters. Based on analysis including the WIO and Madagascar (K = 5, Fig. 2b), four groups can be distinguished (1) sites in Kenya, Tanzania, northern Mozambique and western Madagascar (La, Wa, Di, Mo, Ki, Mi, St, Ja, DS, Mf, Mk, Va, Pe, Na, Nb, Sa, To, An) forming a diverse group of different genetic clusters, although based on cluster distribution, substructures can be identified, (2) sites in southern Mozambique (Vi, Ba, In), (3) north and northeast Madagascar (Ra, Nt, Sm), (4) south of Madagascar (Fa, Lv, FD). Within these groups evidence of mixing of genetic clusters was found at several sample sites: Va, Ja, Ki, Mo (with signature of group 1, but additionally also group 4) and Nb (with signature of group 1, but additionally also 3).
In the PCoA based on pairwise-FST of all sample sites, the first axis divides RM, Mo, Ki, Ja, Va, Ra, Nt, Sm, Fa and Lv from all others. The second axis separates RM, Ki, Ra, Nt and Fa from this segregation, as well as divides La, Wa, Di, Mi, St, DS, Mf, Mk, Na, Pe, Nb, An and FD from Vi, Ba, In, Sa and DS, together explaining 40% of the variation (Fig. 3a). In the PCoA including only sites in the WIO the first axis divides Mo, Ki, Ja, Va, Ra, Nt, Sm, Fa and Lv from all others, and the second axis separates Mo, Ki, Va, Ra and Sm from this segregation, as well as dividing La, Wa, Di, Mi, DS, Mf, Mk, Pe, Na, Ba and In, from St, Vi, Nb, Sa, To, An and FD, together explaining 38% of the variation (Fig. 3b).
Principal coordinates analysis (PCoA) in GenAlEx 6.503 (Peakall and Smouse 2012) based on pairwise FST values (Weir and Cockerham 1984) for Acropora tenuis, the colours representing the Ecoregions as suggested by Obura (2012) see Fig. 1b: a the Red Sea and the Western Indian Ocean, the first two axes explain 40.14% of the variation (the first axis explains 24.79%, the second axis explains 15.35% of the variation); b Western Indian Ocean, the first two axes explain 37.54% of the variation (the first axis explains 25.12%, the second axis explains 12.42% of the variation). For sample site codes see Table 1
In the hierarchical AMOVA, the largest component of variance was found within populations (85%-86% in de WIO; Table 3). Both the grouping based on Marine Ecoregions of the world (Spalding et al. 2007) and the biogeographical ecoregions proposed by Obura (2012), gave identical results in explaining the genetic variance among groupings in the analysis of hierarchical AMOVA in the WIO (component variance among groupings 3%, with FRT = 0.031 for both groupings). Lastly, no significant differences were found in the one-sided comparisons of HO, Ar, FIS and FST between different regions in the WIO.
Discussion
This study describes comprehensive research of the genetic diversity, population structure and connectivity patterns of the broadcast spawning coral Acropora tenuis using microsatellite markers. First the large-scale patterns comparing the Red Sea and the Western Indian Ocean (WIO) are described, followed by the smaller-scale patterns within the WIO and within the southern Mozambique Channel.
Isolation of the Red Sea
The sample site in the Red Sea (RM) was found to be differentiated from all other sites in the WIO, except some sites with low sample size. This confirms the hypothesised genetic break based on Marine Ecoregions of the World, in which the Red Sea and the WIO belong to different provinces (Spalding et al. 2007), which was also recently found for the brooding coral Seriatopora hystrix (van der Ven et al. 2021) and the giant clam Tridacna maxima and T. squamosa (Hui et al. 2016). This isolation can be explained by both historical and contemporary barriers for larval dispersal between the Red Sea and the WIO. The Red Sea was historically isolated from the Indian Ocean by land barriers as a direct result of lower sea levels caused by glacials, and during this time the Red Sea experienced periods of extreme desiccation and hypersalinity (Siddall et al. 2003). Nevertheless, it remains under discussion whether corals survived these conditions in refugia or recolonised the Red Sea from the Gulf of Aden (DiBattista et al. 2016). Contemporary barriers for dispersal between the Red Sea and Indian Ocean are eddies within the central Red Sea (~ 20° N) and limited exchange through the shallow and narrow Bab-el-Mandab Strait. Furthermore, cold-water upwelling in the Gulf of Aden, northern Somalia and Yemen during the summer monsoon, limits the formation of coral habitat in this area (Furnas 2011; Kemp 1998), but see DiBattista et al. (2016). Additionally, the northbound East African Coast Current (EACC) is diverted under the influence of the Somali Counter Current (SC) and larvae are subsequently transported away from the East African coast by the South Equatorial Counter Current (SECC) during the Northeast Monsoon (Gamoyo 2018).
Genetic connectivity in northern Madagascar and the East African mainland
High differentiation was found between the East African mainland and Madagascar and within Madagascar. Similar differentiation between the East African mainland and Madagascar was also found in other coral reef taxa, such as the stony coral S. hystrix (van der Ven et al. 2021), anemonefish Amphiprion akallopisos (Huyghe and Kochzius 2018) and Octopus cyanea (Van Nieuwenhove et al. 2019). Also mangrove taxa showed congruent patterns of differentiation, such as the mangrove tree Rhizophora mucronata (Triest et al. 2021), snails Terebralia palustris (Ratsimbazafy and Kochzius 2018), Littoraria subvittata and L. pallescens (Nehemia et al. 2019), as well as the giant mud crab Scylla serrata (Rumisha et al. 2017).
However, in this study there is evidence for long-distance larval dispersal for A. tenuis in the NMC region, with exchange through the Mozambique Channel between northern Mozambique (Va, Pe, Na) and northern Madagascar (Ra, Nb). This is congruent with the hypothesis that coral reefs within the Northern Mozambique Channel are connected with each other through oceanographic conditions in the channel and share genetic input from the South Equatorial Current (SEC), causing them to have a more similar coral fauna (Obura 2012).
But this larval exchange is not exclusive within the NMC as there is an indication for exchange between site Ra (NMC) and the sites Nt and SM (Eastern Madagascar, EM), which are upstream of the NMC in the Northeast Madagascar Current (NEMC). Also, there is exchange from Nb and Ra with sites downstream of the SEC, such as Va and other sites downstream of the EACC along the coast to Tanzania and Kenya (Mo, Ki, Ja). Long-distance travel of larvae released in northern Mozambique up the coast of Africa (Tanzania, Kenya and up to Somalia) is supported by dispersal models for the WIO for corals with long PLD such as A. tenuis (Crochelet et al. 2016); PLD = 50 days). However, long-distance exchange of larvae is likely infrequent considering the levels of differentiation found in this study based on pairwise FST and DEST values between Mozambique and northern Madagascar, particularly for Ra, while Nb depicts lower levels of differentiation and shows a mixed signature of both mainland sites as well as sites in southwest Madagascar (Sa, To, An). High genetic connectivity was found for most sites along the East African mainland coast, with no indication for strict genetic barriers between the NMC and EAC region. These results reconfirm the high connectivity found for a smaller study on A. tenuis in Tanzania and Kenya (van der Ven et al. 2016) and are congruent with the lack of distinct dispersal barriers found for corals with a substantial lower dispersal capacity (S. hystrix, van der Ven et al. 2021). High connectivity along the coast is supported by biophysical modelling studies, which propose Tanzania as a seeding source of larvae for downstream Kenya (Crochelet et al. 2016; Mayorga-Adame et al. 2016).
Substructure of reefs in the southern Mozambique channel
The sites within the SMC cluster together, although within the SMC there is a substructure based on both clustering and pairwise FST and DEST values between sites in Mozambique (Vi, Ba) and Madagascar (Sa, To, An). This barrier was hypothesised based on the species distribution of corals, for which the SMC is described as a distinct area, but with a potential barrier in the Mozambique Channel (Obura 2012). These results also support a larval dispersal modelling study in the WIO, in which sites in southern Mozambique and southeast Madagascar are isolated from all other sites as well as each other (Crochelet et al. 2016); PLD = 50 days).
The sites in southern Mozambique (Vi, Ba) are significantly differentiated from most other sites in the WIO, except for In (southern Mozambique), which is supported by larval dispersal models that consider this area to be self-recruiting, although connected with sites further south down to South Africa (Crochelet et al. 2016). This differentiation in southern Mozambique can be explained by the isolation of these smaller and patchier reefs that are separated from the northern reefs by an area devoid of coral reefs due to freshwater discharge and sediments from large-scale river outlets in central Mozambique. This pattern is supported by research on the stony coral Platgyra daedalea, in which Bazaruto (in this study Vi) and Inhaca (in this study In) were clustered together and separate from reefs in northern Mozambique (Montoya-Maya et al. 2016), as well as by a studies on Acropora austera, in which panmixia was found between Bazaruto and Inhaca, as well as between some of the Mozambique and South African sites (Macdonald et al. 2011; Montoya-Maya et al. 2016). Also, mangrove taxa, such as the mangrove tree R. mucronata (Triest et al. 2021), as well as the snails L. subvittata (Nehemia et al. 2019) and T. palustris (Ratsimbazafy and Kochzius 2018), show a genetic separation of population in southern Mozambique from other regions in the WIO.
The sites in the southwest of Madagascar show, besides mixing with sites in northern Madagascar (Nb), exchange of larvae with sites in northern Mozambique and Tanzania, as revealed by individual-based cluster analysis. This exchange was also found in drifter experiments (Ramanantsoa et al. 2018), and it supports a larval dispersal modelling study, in which strong southward dispersal and subsequently transport in the Mozambique Channel eddies were found for larvae released from sites south of Pe (Pembe, Mozambique; Gamoyo (2018)). Connectivity between southwest Madagascar and northern Mozambique is also observed in the stony coral S. hystrix (van der Ven et al. 2021). Lastly, southern Madagascar (SM) forms a separate group in Bayesian clustering, but with high differentiation of Lv, potentially caused by eddies and ocean currents directed away from land, high wave action and high asexual reproduction (discussed below).
Inbreeding within all sites and genetic diversity
Significant FIS values indicating heterozygote deficits were found for all sample sites. Heterozygote deficits are common in studies on scleractinian corals and were found in A. tenuis in northern Western Australia (Underwood 2009) and for A. austera in Mozambique and South Africa (Montoya-Maya et al. 2016). Heterozygote deficiencies can be caused by null alleles, but because the deficit was found for all loci in all locations, the contribution of null alleles is unlikely, which is partly supported by the analysis with INEst, indicating that inbreeding rather than null alleles is causing the heterozygote deficits at most sample sites. Additionally, the heterozygote deficits can be attributed to the sampling of different genetic cohorts, also known as the Wahlund effect, or a genetic patchiness caused by biological factors concerning temporal and spatial admixture, which can be amplified by high asexual reproduction (as seen in Fa, Lv, FD, see below). This fits the pattern of long-distance dispersal found in this study and was also found for A. tenuis from northern Western Australia (Underwood 2009).
Asexual reproduction caused by coral fragmentation is common in branching acroporids (Baums et al. 2006; Baums 2008), including A. tenuis (Underwood 2009). High clonal richness was found in three locations in southern Mozambique (Fa, Lv, FD), with 48% to 67% of the colonies formed by clonal fragmentation, compared to an average of 2% in all other sites combined. Since the sampling method in this study was designed to avoid sampling clones, the observed clonal richness might underestimate the total contribution of asexual reproduction in A. tenuis at these sites. The contribution of colonies formed by fragmentation was variable in northern Western Australia, with an average of 10% of sampled colonies for A. tenuis (Underwood 2009). A low contribution of asexual reproduction was also found for A. digitifera in Japan, where 1% of sampled individuals was formed by clones (Nakajima et al. 2010). The reefs on the coast of south Madagascar are influenced by extreme wave action, notably altering the coral's branched growth form towards a more robust and sturdier colony. This wave action is likely also causing fragmentation and subsequently the growth of clonal colonies, which can explain the high clonal richness in this area.
Patterns of genetic connectivity can be used to verify and explain proposed biogeographic regions such as Marine Ecoregions of the World (Spalding et al. (2007) and Ecoregions based on coral diversity (Obura 2012). Here we aimed to infer dispersal patterns to identify potential barriers in the WIO and Red Sea and to verify the proposed Ecoregions. The broadcast spawning coral A. tenuis displays genetic connectivity over long distances within the WIO, but the Red Sea is isolated confirming hypothesis I, which can be attributed to both historical and maintained contemporary barriers for larval dispersal. In the WIO, high genetic connectivity was found in the northern sites along the coast of Kenya and Tanzania and northern part of Mozambique, but sites in the South of Mozambique and offshore sites in the north and south of Madagascar appear less connected. Neither strong genetic barriers, nor differences in genetic diversity could be found within the WIO, rejecting hypothesis II-III based on biogeographic regions as proposed by Obura (2012). However, there is an indication that regions can be defined based on species diversity in Madagascar confirming hypothesis IV. Remarkable is the genetic connectivity found between northeast Madagascar and northern Mozambique and Tanzania, as well as between western Madagascar and the northern Mozambique sites, although this genetic connectivity is probably low or infrequent based on the high differentiation found among these sites. These patterns of high genetic connectivity combined with contemporary dispersal barriers can be explained by the long larval duration of A. tenuis and the prevailing northbound East African Coast Current that facilitates higher genetic connectivity along the northern East African Coast, while eddies in the Mozambique Channel are causing larval retention in south Mozambique and Madagascar.
References
Almany G, Connolly S, Heath D, Hogan J, Jones G, McCook L, Mills M, Pressey R, Williamson D (2009) Connectivity, biodiversity conservation and the design of marine reserve networks for coral reefs. Coral Reefs 28:339–351
Ayre D, Hughes T (2000) Genotypic diversity and gene flow in brooding and spawning corals along the Great Barrier Reef, Australia. Evolution 54:1590–1605
Baird AH, Guest JR, Edwards AJ et al (2021) An Indo-Pacific coral spawning database. Sci Data 8:35. https://doi.org/10.1038/s41597-020-00793-8
Baums IB, Miller MW and Hellberg ME (2006) Geographic variation in clonal structure in a reef‐building Caribbean coral, Acropora palmata. Ecol Monogr 76(4):503–519
Baums IB (2008) A restoration genetics guide for coral reef conservation. Mol ecol 17(12):2796–2811
Besnier F, Glover KA (2013) ParallelStructure: A R package to distribute parallel runs of the population genetics program STRUCTURE on multi-core Computers. Plos One 8:e70651
Bouwmeester J, Baird AH, Chen CJ, Guest JR, Vicentuan KC, Berumen ML (2015) Multi-species spawning synchrony within scleractinian coral assemblages in the Red Sea. Coral Reefs 34:65–77
Burke L, Reytar K, Spalding M, Perry A (2011) Reefs at risk revisited. World Resources Institute, Washington DC
Campagne P, Smouse P, Varouchas G, Silvain J, Leru B (2012) Comparing the van Oosterhout and Chybicki-Burczyk methods of estimating null allele frequencies for inbred populations. Mol Ecol Resour 12:975–982
Chybicki I, Burczyk J (2009) Simultaneous estimation of null alleles and inbreeding coefficients. J Hered 100:106–113
Costanza R, de Groot R, Sutton P, van der Ploeg S, Anderson SJ, Kubiszewski I, Farber S, Turner RK (2014) Changes in the global value of ecosystem services. Global Environ Chang 26:152–158
Cowen R, Sponaugle S (2009) Larval dispersal and marine population connectivity. Annu Rev Mar Sci 1:443–466
Crochelet E, Roberts J, Lagabrielle E, Obura D, Petit M, Chabanet P (2016) A model-based assessment of reef larvae dispersal in the Western Indian Ocean reveals regional connectivity patterns - Potential implications for conservation policies. Reg Stud Mar Sci 7:159–167
DiBattista JD, Roberts MB, Bouwmeester J, Bowen BW, Coker DJ, Lozano-Cortes DF, Howard Choat J, Gaither MR, Hobbs JPA, Khalil MT, Kochzius M, Myers RF, Paulay G, Robitzch VSN, Saenz-Agudelo P, Salas E, Sinclair-Taylor TH, Toonen RJ, Westneat MW, Williams ST, Berumen ML (2016) A review of contemporary patterns of endemism for shallow water reef fauna in the Red Sea. J Biogeogr 43:423–439
Dorken ME, Eckert CG (2001) Severely reduced sexual reproduction in northern populations of a clonal plant, Decodon verticillatus (Lythraceae). J Ecol 89:339–350
Earl D, Vonholdt B (2012) STRUCTURE HARVESTER: a website and program for visualizing STRUCTURE output and implementing the Evanno method. Conserv Genet Resour 4:359–361
Evanno G, Regnaut S, Goudet J (2005) Detecting the number of clusters of individuals using the software STRUCTURE: a simulation study. Mol Ecol 14:2611–2620
Fisher R, O’Leary RA, Low-Choy S, Mengersen K, Knowlton N, Brainard RE, Caley MJ (2015) Species richness on coral reefs and the pursuit of convergent global estimates. Curr Biol 25:500–505
Furnas MJ (2011) Upwelling and Coral Reefs. In: Hopley D. (eds) Encyclopedia of Modern Coral Reefs. Encyclopedia of Earth Sciences Series. Springer, Dordrecht. https://doi.org/10.1007/978-90-481-2639-2
Gamoyo MJ (2018) Modeling dispersal and connectivity of broadcast spawning corals in the Western Indian Ocean. Ph.D. thesis, University of Cape Town.
Gao H, Williamson S, Bustamante CD (2007) A Markov chain Monte Carlo approach for joint inference of population structure and inbreeding rates from multilocus genotype data. Genetics 176:1635–1651
Goudet J (1995) FSTAT (Version 1.2): a computer program to calculate F-statistics. J Her 86:485–486
Graham E, Baird A, Connolly S (2008) Survival dynamics of scleractinian coral larvae and implications for dispersal. Coral Reefs 27:529–539
Hancke L, Roberts MJ, Ternon JF (2014) Surface drifter trajectories highlight flow pathways in the Mozambique Channel. Deep-Sea Research Part Ii-Topical Studies in Oceanography 100:27–37
Hardy OJ, Vekemans X (2002) SPAGEDi: a versatile computer program to analyse spatial genetic structure at the individual or population levels. Mol Ecol Notes 2:618–620
Hoegh-Guldberg O, Poloczanska ES, Skirving W, Dove S (2017) Coral reef ecosystems under climate change and ocean acidification. Front Mar Sci 4:158
Hughes TP, Barnes ML, Bellwood DR, Cinner JE, Cumming GS, Jackson JBC, Kleypas J, van de Leemput IA, Lough JM, Morrison TH, Palumbi SR, van Nes EH, Scheffer M (2017) Coral reefs in the Anthropocene. Nature 546:82–90
Hui M, Kraemer WE, Seidel C, Nuryanto A, Joshi A, Kochzius M (2016) Comparative genetic population structure of three endangered giant clams (Cardiidae: Tridacna species) throughout the Indo-West Pacific: implications for divergence, connectivity and conservation. J Molluscan Stud 82:403–414
Huyghe F, Kochzius M (2018) Sea surface currents and geographic isolation shape the genetic population structure of a coral reef fish in the Indian Ocean. PLoS ONE 13(3): e0193825
Jost L (2008) GST and its relatives do not measure differentiation. Mol Ecol 17:4015–4026
Kemp JM (1998) Zoogeography of the coral reef fishes of the Socotra Archipelago. J Biogeogr 25:919–933
Lutjeharms J, Bornman T (2010) The importance of the greater Agulhas Current is increasingly being recognised. S Afr J Sc 106:1–4
Macdonald A, Schleyer M, Lamb J (2011) Acropora austera connectivity in the south-western Indian Ocean assessed using nuclear intron sequence data. Mar Biol 158:613–621
Mangubhai S, Harrison PL (2008) Asynchronous coral spawning patterns on equatorial reefs in Kenya. Mar Ecol Prog Ser 360:85–96
Mangubhai S, Harrison PL (2009) Extended breeding seasons and asynchronous spawning among equatorial reef corals in Kenya. Mar Ecol Prog Ser 374:305–310
Mayorga-Adame CG, Strub PT, Batchelder HP, Spitz YH (2016) Characterizing the circulation off the Kenyan-Tanzanian coast using an ocean model. J Geophys Res Oceans 121:1377–1399
McClanahan TR, Ateweberhan M, Darling ES, Graham NAJ, Muthiga NA (2014) Biogeography and change among regional coral communities across the Western Indian Ocean. Plos One 9:e93385
Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, Passarotti M, Kaufman S, O’Leary MA (2015) A RESTful API for Access to Phylogenetic Tools via the CIPRES Science Gateway. Evol Bioinform 11:43–48
Moberg F, Folke C (1999) Ecological goods and services of coral reef ecosystems. Ecol Econ 29:215–233
Montoya-Maya PH, Schleyer MH, Macdonald AHH (2016) Limited ecologically relevant genetic connectivity in the south-east African coral populations calls for reef-level management. Mar Biol 163:171
Nakajima Y, Nishikawa A, Iguchi A, Sakai K (2010) Gene flow and genetic diversity of a broadcast-spawning coral in northern peripheral populations. Plos One 5:e11149
Nehemia A, Ngendu Y, Kochzius M (2019) Genetic population structure of the mangrove snails Littoraria subvittata and L. pallescens in the Western Indian Ocean. J Exp Mar Biol Ecol 514–515:27–33
Nishikawa A, Sakai K (2005) Settlement-competency period of planulae and genetic differentiation of the scleractinian coral Acropora digitifera. Zool Sci 22:391–399
Nishikawa A, Katoh M, Sakai K (2003) Larval settlement rates and gene flow of broadcast-spawning (Acropora tenuis) and planula-brooding (Stylophora pistillata) corals. Mar Ecol Prog Ser 256:87–97
Obura D (2012) The diversity and biogeography of Western Indian Ocean reef-building corals. Plos One 7:e45013
Obura D, Gudka M, Rabi FA, Gian SB, Bijoux J, Freed S, Maharavo J, Mwaura J, Porter S, Sola E, Wickel J, Yahya S, Ahamada S (2017) Coral reef status report for the Western Indian Ocean. Global Coral Reef Monitoring Network (GCRMN)/International Coral Reef Initiative (ICRI)
Palumbi S (2003) Population genetics, demographic connectivity, and the design of marine reserves. Ecol Appl 13:146–158
Peakall R, Smouse P (2012) GenAlEx 6.5: genetic analysis in Excel. Population genetic software for teaching and research-an update. Bioinformatics 28:2537–2539
Pritchard J, Stephens M, Donnelly P (2000) Inference of population structure using multilocus genotype data. Genetics 155:945–959
Ramanantsoa JD, Penven P, Krug M, Gula J, Rouault M (2018) Uncovering a new current: The Southwest Madagascar Coastal Current. Geophys Res Lett 45:1930–1938
Ratsimbazafy HA, Kochzius M (2018) Restricted gene flow among Western Indian Ocean populations of the mangrove whelk Terebralia palustris (Linnaeus, 1767) (Caenogastropoda: Potamididae). J Molluscan Stud 84:163–169
Richards ZT, van Oppen MJH (2012) Rarity and genetic diversity in Indo-Pacific Acropora corals. Ecol Evol 2(8):1867–1888
Rumisha C, Huyghe F, Rapanoel D, Mascaux N, Kochzius M (2017) Genetic diversity and connectivity in the East African giant mud crab Scylla serrata: Implications for fisheries management. PLoS ONE 12(10): e0186817
Schott F, McCreary J (2001) The monsoon circulation of the Indian Ocean. Prog Oceanogr 51:1–123
Siddall M, Rohling EJ, Almogi-Labin A, Hemleben C, Meischner D, Schmelzer I, Smeed DA (2003) Sea-level fluctuations during the last glacial cycle. Nature 423:853–858
Sofianos SS, Johns WE (2003) An Oceanic General Circulation Model (OGCM) investigation of the Red Sea circulation: 2. Three-dimensional circulation in the Red Sea. J Geophys Res Oceans 108:15
Sola E, da Silva IM, Glassom D (2016) Reproductive synchrony in a diverse Acropora assemblage, Vamizi Island, Mozambique. Mar Ecol-Evol Persp 37:1373–1385
Spalding MD, Fox HE, Halpern BS, McManus MA, Molnar J, Allen GR, Davidson N, Jorge ZA, Lombana AL, Lourie SA, Martin KD, McManus E, Recchia CA, Robertson J (2007) Marine ecoregions of the world: A bioregionalization of coastal and shelf areas. Bioscience 57:573–583
Triest L, Van der Stocken T, De Ryck D, Kochzius M, Lorent S, Ngeve M, Ratsimbazafy HA, Sierens T, van der Ven R, Koedam N (2021) Expansion of the mangrove species Rhizophora mucronata in the Western Indian Ocean launched contrasting genetic patterns. Sci Rep 11:4987. https://doi.org/10.1038/s41598-021-84304-8
Underwood JN (2009) Genetic diversity and divergence among coastal and offshore reefs in a hard coral depend on geographic discontinuity and oceanic currents. Evol Appl 2:222–233
Underwood JN, Smith LD, van Oppen MJH, Gilmour JP (2009) Ecologically relevant dispersal of corals on isolated reefs: implications for managing resilience. Ecol Appl 19:18–29
van der Ven RM, Triest L, De Ryck DJR, Mwaura JM, Mohammed MS, Kochzius M (2016) Population genetic structure of the stony coral Acropora tenuis shows high but variable connectivity in East Africa. J Biogeogr 43:510–519
van der Ven RM, Flot J-F, Buitrago-López C, Kochzius M (2021) Population genetics of the brooding coral Seriatopora hystrix reveals patterns of strong genetic differentiation in the Western Indian Ocean. Heredity 126(2):351–365
Van Nieuwenhove AHM, Ratsimbazafy HA, Kochzius M (2019) Cryptic diversity and limited connectivity in octopuses: recommendations for fisheries management. PLoS One 14(5):e0214748
Veron JEN (2000) Corals of the world. Australian Institute of Marine science, Townsville
Waits LP, Luikart G, Taberlet P (2001) Estimating the probability of identity among genotypes in natural populations: cautions and guidelines. Mol Ecol 10:249–256
Weersing and Toonen (2009) Population genetics, larval dispersal, and connectivity in marine systems. MEPS 436:291–305
Weir BS, Cockerham CC (1984) Estimating F-statistics for the analysis of population-structure. Evolution 38:1358–1370
Wilkinson C (2002) Status of coral reefs of the world, Coral Reef Monitoring Network, Townsville
Acknowledgements
We would like to thank the funding agencies that supported our work: Fonds Wetenschappelijk Onderzoek Vlaanderen (FWO) for funding the project ‘Connectivity of Coral Reefs and Mangroves in the Western Indian Ocean’ (COCOMA-WIO; grant 1501612N); Vrije Universiteit Brussel (VUB, OZR2068BOF) for financial support; Vlaamse Interuniversitaire—Raad Universitaire Ontwikkelingssamenwerking (VLIR-UOS) for providing a PhD scholarship to H.A. Ratsimbazafy; VLIR-UOS, doctoral school of the VUB, and Leopold III-fonds for providing travel grants to R.M. van der Ven; European Union Erasmus Mundus Programme CARIBU, VLIR-UOS and King Abdullah University for Science and Technology (Saudi Arabia) for providing travel grants to M. Kochzius; T. Sierens (VUB, Belgium) for assistance during laboratory work; L. van Dijk for processing samples; T. Lavitra (Institut Halieutiques et des Sciences Marine, Université de Tuléar, Madagascar), J. M. Mwaura (Kenya Marine and Fisheries Research Institute, Kenya) and B. Cowburn (University of Oxford, UK) for collecting samples; D. De Ryck (VUB, Belgium), F. Huyghe (VUB, Belgium), M.S. Mohammed (State University of Zanzibar, Tanzania), V. Muhala (Eduardo Mondlane University, Mozambique) and M. Berumen (King Abdullah University for Science and Technology) for assistance during fieldwork; M. Sheikh (State University of Zanzibar) for logistical support; the competent authorities in Kenya, Madagascar, Mozambique, Saudi Arabia and Tanzania for permits; N. van Hoytema (Centre for Environment, Fisheries and Aquaculture Science (Cefas), UK) for proofreading.
Funding
Fonds Wetenschappelijk Onderzoek, COCOMA-WIO, grant 1501612 N, Vrije Universiteit Brussel, VUB, OZR2068BOF, VLIR-UOS, Leopold III-fonds, European Union Erasmus Mundus Programme CARIBU.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Topic Editor Danwei Huang
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
van der Ven, R.M., Ratsimbazafy, H.A. & Kochzius, M. Large-scale biogeographic patterns are reflected in the genetic structure of a broadcast spawning stony coral. Coral Reefs 41, 611–624 (2022). https://doi.org/10.1007/s00338-022-02220-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-022-02220-3