Abstract
Current seawater temperatures around the northeastern Arabian Peninsula resemble future global forecasts as temperatures > 35 °C are commonly observed in summer. To provide a more fundamental aim of understanding the structure of wild populations in extreme environmental conditions, we conducted a population genetic study of a widespread, regional endemic table coral species, Acropora downingi, across the northeastern Arabian Peninsula. A total of 63 samples were collected in the southern Arabian/Persian Gulf (Abu Dhabi and Qatar) and the Sea of Oman (northeastern Oman). Using RAD-seq techniques, we described the population structure of A. downingi across the study area. Pairwise G’st and distance-based analyses using neutral markers displayed two distinct genetic clusters: one represented by Arabian/Persian Gulf individuals, and the other by Sea of Oman individuals. Nevertheless, a model-based method applied to the genetic data suggested a panmictic population encompassing both seas. Hypotheses to explain the distinctiveness of phylogeographic subregions in the northeastern Arabian Peninsula rely on either (1) bottleneck events due to successive mass coral bleaching, (2) recent founder effect, (3) ecological speciation due to the large spatial gradients in physical conditions, or (4) the combination of seascape features, ocean circulation and larval traits. Neutral markers indicated a slightly structured population of A. downingi, which exclude the ecological speciation hypothesis. Future studies across a broader range of organisms are required to furnish evidence for existing hypotheses explaining a population structure observed in the study area. Though this is the most thermally tolerant acroporid species worldwide, A. downingi corals in the Arabian/Persian Gulf have undergone major mortality events over the past three decades. Therefore, the present genetic study has important implications for understanding patterns and processes of differentiation in this group, whose populations may be pushed to extinction as the Arabian/Persian Gulf warms.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
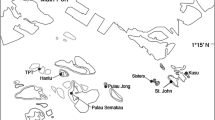
The northeastern Arabian Peninsula, i.e., the sea area bounded by the Arabian/Persian Gulf (hereafter called ‘the Gulf’) and the Sea of Oman (Fig. 1), is distinguished by a remarkable shift in both spatial and temporal oceanographic conditions (Sheppard 1992). The Gulf, which extends from southern Iraq to the Strait of Hormuz, is a shallow (mean depth 36 m) semi-enclosed body of water surrounded by hyper-arid land. High evaporation rates and restricted water exchange with the open ocean create extreme environmental conditions (Kämpf and Sadrinasab 2006). This extreme marine environment is characterized by dense, hypersaline water (often > 42 ppt) (Swift and Bower 2003), and with summer maximum sea surface temperatures (SST) exceeding 35 °C, making the Gulf the warmest sea on Earth (Vaughan et al. 2019). During the winter, Gulf SSTs fall well below values normally associated with tropical marine ecosystems, reaching minima < 13 °C (Coles 2003).
Map of the northeastern Arabian Peninsula and sampling sites. The color background represents the annual mean SST. Font: www.bio-oracle.org
By contrast, the Sea of Oman is a deep oceanic basin (> 2000 m depth) exhibiting more moderate physical conditions compared to the Gulf. In this basin, SSTs generally reach only 30 °C during the summer, whereas in winter, SSTs generally remain above 23 °C (Claereboudt 2019). Both seas are connected by the narrow Strait of Hormuz (56 km wide at its narrowest point), through which the Gulf receives surface driven input of low-salinity Indian Ocean Surface Water (IOSW), replacing water lost by evaporation within the Gulf (Vaughan et al. 2019). On the other hand, the Sea of Oman receives Persian Gulf Water (PGW) that is characterized by high salinity and temperatures, and which exits the Gulf along the seafloor due to elevated salinity-driven density (Swift and Bower 2003; Claereboudt 2019).
The extreme environmental conditions of the Gulf exert considerable impact on the abundance and distribution of marine biota (Sheppard 1993; Wilson et al. 2002). Corals and reef-associated fauna in this basin generally display low diversity compared to other seas bordering the Arabian Peninsula (Wilson et al. 2002; Coles 2003; Burt et al. 2011; Hoey et al. 2016; Bouwmeester et al. 2021). For example, out of the 401 scleractinian coral species recorded in waters surrounding the Peninsula, the Gulf contains 66 species (Riegl et al. 2012; DiBattista et al. 2016a; Berumen et al. 2019), versus over 100 species in the Sea of Oman (Claereboudt 2019) and 359 species in the Red Sea (Berumen et al. 2019). Thus, biogeographic studies suggest that the Gulf is unique in terms of marine community composition (DiBattista et al. 2016a), with communities that are constrained by their environmental tolerance (Sheppard and Sheppard 1991; Coles 2003).
The extreme summer temperatures currently encountered by reef fauna in the Gulf are projected to become widespread across the tropics in the coming decades (Bauman et al. 2011; Bouwmeester et al. 2021). As such, climate change represents a substantial threat to marine biodiversity globally, where reef fauna have evolved in relatively stable thermal environments (Leadley et al. 2010). Hence, there is growing research on corals that currently exist in high-temperature environments, to better understand how reef organisms may adapt and respond to potential scenarios of near future climate change (Burt et al. 2020). Much of this interest has focused on the northeastern Arabian Peninsula (Burt 2013; Vaughan and Burt 2016; Ben-Hasan and Christensen 2019; Howells et al. 2020), where today’s extreme environmental conditions represent a ‘natural laboratory’ for future conditions elsewhere, and can thus support an understanding of the patterns and processes governing life at extreme temperatures.
The continued persistence of corals and their dependent fauna in the Gulf indicates the potential of these organisms to biologically adapt to one of the most stressful physical environments encountered by scleractinian corals (Sheppard et al. 1992). The genetic differentiation of conspecifics across thermal gradients and the geographical variation in bleaching susceptibility (Osman et al. 2018) indicate that the ability of corals to survive under extreme conditions may be due to their genetic adaptation (Wilson et al. 2002; Liew et al. 2020). For example, in the northeastern Arabian Peninsula, the colonies of both Platygyra daedalea (Howells et al. 2016) and their heat-tolerant algal symbiont Cladocopium thermophilum (Smith et al. 2017) display distinct genetic clusters between the Gulf and the Sea of Oman. Furthermore, P. daedalea coral larvae from the Gulf are significantly more thermally tolerant than conspecific larvae from the Sea of Oman (Howells et al. 2016). Thus, in the context of metapopulation dynamics under global warming scenarios, corals inhabiting already warming regions (e.g., the Gulf) might avoid local population declines in adjacent areas (e.g., Sea of Oman) by spreading existing heat tolerance alleles via larval migration (Dixon et al. 2015).
Alternatively, it has been suggested that, at least in the short-term, physiological plasticity in thermal tolerance will play a key role in coral response to climate change (Palumbi et al. 2014). The Gulf is a very young sea that began to form less than 15,000 yrs ago, having been completely dry during the late Pleistocene and refilled only at the beginning of the Holocene (Sarnthein 1972; Sheppard 1993; Lokier et al. 2015). Marine water likely transgressed an average of > 100 m per year (Teller et al. 2000) and the modern shorelines developed ca. 3000–6000 yrs ago (Lambeck 1996; Vaughan et al. 2019), which indicates that coral species were established in the Gulf just in the last few millennia. More recently, mass coral bleaching and mortality of > 80% of corals occurred four times in recent decades, first in the late 1970s, then again in 1996, 1998, and 2017, which significantly reduced live coral cover in the Gulf, particularly Acropora species (Riegl et al. 2018; Burt et al. 2019; Paparella et al. 2019). Therefore, given the recent foundation and the consecutive bottleneck scenarios in the Gulf, the hypothesis of genetic adaptability is valid only if strong selection is assumed so that Acropora species could rapidly adapt to the extreme conditions during only the last few millennia (Torda et al. 2017). In contrast, the reduced population size of Acropora species during consecutive bottlenecks in the Gulf creates better conditions for the action of genetic drift rather than natural selection.
Here, we hypothesize that Acropora downingi, a widespread, regional endemic table coral species, displays a genetic discontinuity across the northeastern Arabian Peninsula. This hypothesis emerges from existing genomics data for other coral species and biophysical models of larval dispersal. The former showed that the genetic discontinuity of P. daedalea populations and their zooxanthellae symbionts coincides geographically with the Strait of Hormuz (Howells et al. 2016; Hume et al. 2018). The latter, in turn, revealed predominance of self-recruitment within the Gulf (Cavalcante et al. 2020), and that water exchange through the strait may act as a hard physical barrier to larval dispersal (and hence gene flow) in scenarios of high mortality rate and short pelagic larval duration (Torquato and Møller 2020). Therefore, our hypothesis assumes that rather than natural selection, genetic differentiation in A. downingi is due to genetic drift following either recent foundation or successive bottlenecks, imposed by mass bleaching events. In the present study, we used Restriction site Associated DNA Sequencing (RAD-Seq) to test this hypothesis, by using genome-wide neutral markers (1) to describe and compare the patterns of genetic diversity distribution of a regional endemic coral species from reef sites in the northeastern Arabian Peninsula, and (2) to investigate whether sufficient generations since the foundation or the bottlenecks have passed in order to observe divergence in A. downingi populations due to the action of genetic drift.
Material and methods
Sampling design and DNA extraction
Acropora downingi were collected (n = 63) between March 2017 and November 2017 from 11 sites spanning three sampling regions: Abu Dhabi and Qatar in the southern Gulf, and the northern Sea of Oman (Fig. 1, Table 1 and Supplementary Table S1). The coral samples were identified following Riegl et al. (2012), Wallace (1999) and Wallace et al. (2012), and promptly preserved in 96–99% EtOH and kept frozen at − 20 °C until DNA extraction. Underwater images were taken from each sampled colony to collect habitat, growth form and close-up polyp information. Reference material collected from each sampled site is also available from Qatar and is stored at Qatar University. High-molecular-weight genomic DNA was isolated in a final elution volume of 100 μl following the manufacturer’s instructions for Qiagen DNeasy blood and tissue kit. Subsequently, the extract concentrations were measured using the Qubit 2.0 dsDNA BR Assay Kit (IntrogenTM) Fluorometer and checked for molecular weight bands on a 1% agarose gel.
Genotyping and de novo assembly of RAD tags
RAD tag libraries were prepared by Floragenex Inc following the protocol depicted in Baird et al. (2008). In short, the genomic DNA was single-digested utilizing the restriction enzyme PstI and ligated to in-line barcode sequences that allowed the resulting amplified fragments. Restriction fragments representing a reduced part of the original genome from all individuals, were then pooled (multiplexed), randomly sheared and size‐selected (300–500 bp, average of ~ 380 bp). Finally, the DNA fragments were PCR amplified and the RAD-seq libraries single-read (1 × 100 bp) sequenced on two lanes using the Illumina HiSeq2000 platform (see Davey and Blaxter 2010; Etter and Johnson 2011).
Raw reads obtained from 100 bp single-end Illumina sequencing were assessed for sequence quality, AT/GC content, and duplicate or overrepresented sequences using FastQC v.0.11.5. After initial quality assessment, reads were filtered and detection of single nucleotide polymorphism (SNP) was performed in Stacks v.2.2 pipeline (Catchen et al. 2011, 2013).
First, we used the process_radtags program for removal of low-quality reads and demultiplexing, and the remainder sequences were trimmed to 90 base pairs. Secondly, as a reference genome was not available for A. downingi, RAD tags were analyzed de novo with parameters optimized using the r80 method outlined in Paris et al. (2017) and thoroughly described in Rochette and Catchen (2017). Thus, de novo assemble exhibited the maximum number of polymorphic loci shared across 80% of the individuals when the parameters were M = 5, m = 3, n = 4. Potential contaminants (i.e., bacteria, Symbiodiniaceae spp. and viruses) were identified from a FASTA file, containing all individuals, using BLASTn algorithm (E-value of 1e−10; Supplementary Table S2). Next, a blacklist in Stacks' populations program was used to remove loci that matched contaminants sequences. In the populations program RAD tags for all individuals were used to detect only the first SNP (–write_single_snp) by identifying at the same locus a marker that was present in one set of individuals but absent in another.
Second filtering procedures and outlier loci
All second filtering procedures were performed in Plink v.1.9, and excluded individuals with more than 10% missing genotypes, and only SNPs with a 90% genotyping rate (10% missing) and a minor allele frequency (MAF) higher than 5% were maintained. Furthermore, markers that did not meet the Hardy–Weinberg Equilibrium assumptions were also excluded.
Furthermore, we scanned our data to identify a list of outlier loci by using a Bayesian approach implemented in BayeScan (Foll and Gaggiotti 2008). To do so, Plink files were converted to the BayeScan format using PGDSpider v 2.1.1.5 software (Lischer and Excoffier 2012). The outlier loci analysis was then run with a prior odds ratio of 10 (indicating that for a given SNP the neutral model is 10 times more likely than the model with selection), and all other default settings, using a q-value cut-off of 0.05 to determine outliers.
Summary statistic and population genetic structure
Pairwise differentiation (G’st) and genetic diversity metrics (see Nei 1987) were estimated using GENODIVE version 2.0 (Meirmans and Van Tienderen 2004). We estimated the observed heterozygosity (Ho) that ranges from 0 (all individuals are homozygous) to 1 (all individuals are heterozygous), the heterozygosity within Qatar, Abu Dhabi and Oman samples (Hs) and the total heterozygosity (Ht). The significance of pairwise G’st values was tested using 1000 permutations.
Distance‐based method and model-based clustering analyses were used to estimate population genetic structure. The first was performed using a Principal Component Analysis (PCA) with the EIGENSOFT v.6.1.3 program to identify patterns in the data, by highlighting similarities and differences between samples and sampling sites. For the model-based analysis, the number of genetic clusters and the membership of each individual to these clusters were estimated using the ADMIXTURE software (Alexander et al. 2009). Here, the most likely number of clusters was selected based on cross-validation error (CV) and the lowest value of K was chosen (Alexander and Lange 2011). The prior expectation for the possible range of K (from 1 to 3) was based on the number of regions from which the samples were collected, i.e., Qatar, Abu Dhabi and Oman.
Results
RAD-seq summary
The RAD library of the two lanes yielded a total of 510,227,840 reads obtained for 63 individual samples (4,049,427.3 ± 2,551,382.2; mean ± SD) from the 11 sites. After process_radtag quality filtering and demultiplexing, 502,054,112 reads remained, comprising 98.4% of all reads (Supplementary Table S3). In denovo_map.pl, 395,628,079 reads were retained and assembled into a total of 9,312,616 loci (147,819.3 ± 65,011.5; mean ± SD) and 1,142,563 (18,135.9 ± 9357.8; mean ± SD) putative allele, with the average depth of coverage per individual ranged between 19 and 82X (36.76X ± 14.29; mean ± SD) (Supplementary Table S4). Finally, in the populations module, a total of 5729 SNPs were obtained.
Additional filtering performed in Plink, such as missing genotype rate, minor allele frequency, linkage disequilibrium and nonconformance with Hardy–Weinberg Equilibrium, excluded one individual from Qatar (Gulf) and one individual from the Sea of Oman due to missing genotype data. From the 1420 SNPs found after the second filtering, Bayescan detected 1 significant locus after FDR correction. Finally, a total of 1419 genome-wide SNPs from 61 individuals were assumed to not be significantly under selection, and thereby they were used in downstream analyses.
Genetic diversity and pairwise differentiation
A summary of the principal statistics, i.e., Hs, Ho and Ht are presented in Table 2, and pairwise differentiation in Table 3. The Qatar individuals displayed the highest heterozygosity. Pairwise G’st showed a significant difference between the Gulf regions and the Sea of Oman. Likewise, when the populations were grouped according to clustering results (see below), a global G’st of 0.062 and pairwise genetic distance of 0.066 was exhibited between the southern Gulf versus the Sea of Oman.
Population genetic structure
The top two principal components (PCs) from PCA analysis explained 8.77% (PC1—5.35% and PC2—3.42%) of the total of genotypic variation, and distinguished individuals sampled in the southern Gulf from those obtained in the western Sea of Oman (Fig. 2). In turn, the Admixture analysis did not indicate the presence of population structure within the dataset. Cross-validation error (CV) determined K = 1 (CV = 0.62449) as the most likely number of clusters for A. downingi. However, this was only slightly more likely than K = 2 (CV = 0.66055), which partitioned individuals from the southern Gulf and western Sea of Oman into two distinct groups (Fig. 3).
Discussion
The aims of this study were to (1) describe the distribution of genetic diversity of Acropora downingi across the northeastern Arabian Peninsula, and (2) test for the hypothesis of neutral genetic differentiation between the world’s warmest sea (i.e., The Gulf) and the adjacent Sea of Oman. Our results suggest a slightly structured population. The distance-based approach used in our investigation suggested a genetic discontinuity within the northeastern Arabian phylogeographic province, with coral populations falling into two distinct subpopulations: the Gulf versus the Sea of Oman. Prior to this paper, a similar study using the RAD-seq approach also indicated a genetic discontinuity for the coral Platygyra daedalea between both seas (Howells et al. 2016). These results suggest that the Gulf represents a distinct phylogeographic unit for coral species (Howells et al. 2016), though not for coral-dependent fishes (Priest et al. 2016; Torquato et al. 2019), likely explained by the substantially longer larval pelagic duration for the latter (Kinlan and Gaines 2003; Shanks et al. 2003). Investigations using microsatellites on both ray-finned fish (van Herwerden et al. 2006) and four shark species (Spaet et al. 2015) did not indicate any discontinuity across the northeastern Arabian Peninsula, though these species exhibit extensive adult movements and are not coral-dependent.
Nevertheless, data on lineage distribution alone do not provide convincing proof that explains the processes underlying this differentiation. Hypotheses to explain distinctiveness of phylo- and biogeography subregions around the Arabian Peninsula have relied on allopatric, parapatric or sympatric geographic patterns. Here, we will not discuss the sympatric mode. The hypothesis supported in our investigation assumes that the observed genetic differentiation in the study area relies on the neutral theory of molecular evolution (Kimura 1983). For this to happen the fixation of neutral allele by genetic drift mainly occurs in scenarios of small population size (e.g., due to bottlenecks) and where the gene flow is constrained.
The reduction of the gene flow is attributed to the combination of seascape features, ocean circulation and larval traits. This argument assumes that the Strait of Hormuz acts as a physical barrier to larval dispersal (and gene flow), by constraining larval movements between the Gulf and the Sea of Oman. Specifically, biophysical models simulating larval transport around the Arabian Peninsula have shown that the connectivity through the Strait of Hormuz was not symmetric; and in the most extreme results, propagules released from the Sea of Oman were not able to cross this barrier if they drifted for 20 days and exhibited high mortality rate (Torquato and Møller 2020). While that model was used to investigate fish larvae, these conditions are in accordance with experiments with Acropora showing that larval survival declined sharply during the first few weeks after spawning compared to other coral species, with the 50% survival point reached within 4.25 days (Graham et al. 2008). Moreover, pelagic larval duration can be further constrained in warmer waters, reducing the spatial scale of connectivity (Munday et al. 2009), and hence promote genetic differentiation between adjacent populations.
In the Arabian Peninsula, the allopatric pattern has been used to explain, for example, the difference of both vertebrate and invertebrate species composition between the Gulf of Aden and the adjacent Red Sea. The formation of an extrinsic barrier to gene flow occurred repeatedly, caused by lowering of sea level during the Pliocene and Pleistocene (4–3 Ma; DiBattista et al. 2016b), such that water flow between both seas through the Bab-el-Mandeb strait was largely interrupted. Unlike the Red Sea, the Gulf is a young basin whose modern shorelines were not formed until 3000–6000 yrs ago (Lambeck 1996; Vaughan et al. 2019), and hence Pleistocene vicariance events have not occurred between the Gulf and the adjacent Sea of Oman.
The parapatric argument, in turn, relies on ecological processes driven by the large spatial gradients and temporal fluctuations of physical conditions across the peninsula (Nanninga et al. 2014; Giles et al. 2015). This process is very sensitive to parameters, such as the migration rate, intensity of selection for local adaptation, population size, and mutation rate (Gavrilets 2004). Nevertheless, despite the potential for strong selection given the extreme environment conditions and the reduced gene flow due to constrained dispersal through the Strait of Hormuz, endemic coral species have not been recorded for the Gulf.
Furthermore, the parapatric speciation-selection hypothesis is also constrained due to small population sizes of Acropora species because of the large and recurrent mortality events within the Gulf (Riegl and Purkis 2015; Riegl et al. 2018). The smaller the population size, the more important is the effect of genetic drift, whereas the effect of natural selection only overwhelms that of drift in the context of large population size. Therefore, genetic drift can have major effects when a population is sharply reduced in size by a natural disaster, such as the successive bottlenecks recorded for Acropora species in the Gulf (Riegl et al. 2018). In addition to coral bleaching and subsequent coral mortality, the reduction of coral populations within the Gulf is also related to both low availability of suitable substratum and the loss of habitat due to anthropogenic factors, such as ongoing urbanization (Sheppard 2016; Burt and Bartholomew 2019).
While the population of A. downingi in the Gulf is slightly distinct from the Sea of Oman population, its future is uncertain. Acropora corals in the Gulf have undergone major mortality events in the past three decades, often with losses of over 90% cover, with populations currently at the brink of collapse (Riegl et al. 2018). Alarmingly, juveniles of Acropora are mostly absent from the coral community (Bento et al. 2017; Pratchett et al. 2017), reducing possibilities of recovery. Acropora downingi has now largely been extirpated from shallow and nearshore environments in the southern Gulf, and only occurs around offshore islands and seamounts where temperatures are buffered by deeper surrounding waters (Burt et al. 2013, 2016, 2019).
Future perspectives
Rapid advances in molecular genetics are now opening avenues in the analysis of adaptive speciation. To test the selection hypothesis, future genetic studies, particularly those using advanced genomic approaches (e.g., whole genome sequencing), could characterize regions exhibiting potential signatures of selection across the entire genome of coral species inhabiting the Gulf. In addition, the results depicted here suggest further investigation onto the neutral hypothesis to distinguish between patterns caused owing to both founder effect and bottleneck events. Therefore, we recommend future studies on demographic events that produced the present-day patterns of genetic diversity, such as changes in population sizes, geographic range expansions, and varying levels of gene flow. For that to happen, reference genomes for coral species are required, as well as additional sampling sites spread out throughout the study area in order to provide greater resolution for the role of the Gulf in the persistence of both present-day and future marine metapopulations.
In addition, the molecular results presented in here, as well as in previous studies, provide incredible value for science and conservation, as they indicate that despite being a young marine habitat the Gulf harbors distinct genetic lineages not found anywhere else. Nevertheless, the extreme environmental and human activities (e.g., dredging and land reclamation) continue causing damage to corals throughout the region. Thus, the corals in our study area are now at risk of being not only locally extirpated, but pushed to extinction as the Gulf warms, and hence conservation actions for this group are urgent.
Data availability
The genetic data reported in this paper have been deposited in the National Center for Biotechnology Information (NCBI) Short Read Archive under the following BioProject Accession Numbers: PRJNA750412 (Acropora downingi in northeastern Arabian Peninsula).
References
Alexander DH, Lange K (2011) Enhancements to the ADMIXTURE algorithm for individual ancestry estimation. BMC Bioinformatics 12:1–6
Alexander DH, Novembre J, Lange K (2009) Fast model-based estimation of ancestry in unrelated individuals. Genome Res 19:1655–1664
Baird NA, Etter PD, Atwood TS, Currey MC, Shiver AL, Lewis ZA, Selker EU, Cresko WA, Johnson EA (2008) Rapid SNP discovery and genetic mapping using sequenced RAD markers. PloS one 3:e3376
Bauman AG, Baird AH, Cavalcante GH (2011) Coral reproduction in the world’s warmest reefs: southern Persian Gulf (Dubai, United Arab Emirates). Coral Reefs 30:405–413
Ben-Hasan A, Christensen V (2019) Vulnerability of the marine ecosystem to climate change impacts in the Arabian Gulf - an urgent need for more research. Glob Ecol Conserv 17:e00556
Bento R, Feary DA, Hoey AS, Burt JA (2017) Settlement patterns of corals and other benthos on reefs with divergent environments and disturbances histories around the northeastern Arabian peninsula. Front Mar Sci 4:305
Berumen ML, Arrigoni R, Bouwmeester J, Terraneo TI, Benzoni F (2019) Corals of the Red Sea. In: Voolstra CR, Berumen ML (eds) Coral Reefs of the Red Sea. Springer, Cham, pp 123–155
Bouwmeester J, Riera R, Range P, Ben-Hamadou R, Samimi-Namin K, Burt JA (2021) Coral and reef fish communities in the thermally extreme Persian/Arabian Gulf: insights into potential climate change effects. In: Rossi S, Bramanti L (eds) Perspectives on the marine animal forest of the world. Springer, Cham, pp 63–86
Burt JA (2013) The growth of coral reef science in the Gulf: A historical perspective. Mar Pollut Bull 72:289–301
Burt JA, Bartholomew A (2019) Towards more sustainable coastal development in the Arabian Gulf: Opportunities for ecological engineering in an urbanized seascape. Mar Pollut Bull 142:93–102
Burt JA, Feary DA, Bauman AG, Usseglio P, Cavalcante GH, Sale PF (2011) Biogeographic patterns of reef fish community structure in the northeastern Arabian Peninsula. ICES J Mar Sci 68:1875–1883
Burt J, Al-Khalifa K, Khalaf E, AlShuwaik B, Abdulwahab A (2013) The continuing decline of coral reefs in Bahrain. Mar Pollut Bull 72:357–363
Burt JA, Smith EG, Warren C, Dupont J (2016) An assessment of Qatar’s coral communities in a regional context. Mar Pollut Bull 105:473–479
Burt JA, Paparella F, Al-Mansoori N, Al-Mansoori A, Al-Jailani H (2019) Causes and consequences of the 2017 coral bleaching event in the southern Persian/Arabian Gulf. Coral Reefs 38:567–589
Burt JA, Camp EF, Enochs IC, Johansen JL, Morgan KM, Riegl B, Hoey AS (2020) Insights from extreme coral reefs in a changing world. Coral Reefs 39:495–507
Catchen J, Hohenlohe PA, Bassham S, Amores A, Cresko WA (2013) Stacks: an analysis tool set for population genomics. Mol Ecol 22:3124–3140
Catchen JM, Amores A, Hohenlohe P, Cresko W, Postlethwait JH (2011) Stacks: building and genotyping loci de novo from short-read sequences. G3-Genes Genom Genet 1:171–182
Cavalcante G, Vieira F, Mortensen J, Ben-Hamadou R, Range P, Goergen E, Campos E, Riegl B (2020) Biophysical model of coral population connectivity in the Arabian/Persian Gulf. Mar Biol 87:193
Claereboudt MR (2019) Oman. In: Sheppard C (ed) World seas: An environmental evaluation. Academic Press, Massachuetts, pp 25–34
Coles S (2003) Coral species diversity and environmental factors in the Arabian Gulf and the Gulf of Oman: a comparison to the Indo-Pacific region. Atoll Res Bull 507:1–19
Davey JW, Blaxter ML (2010) RADSeq: next-generation population genetics. Brief Funct Genom 9:416–423
DiBattista JD, Choat JH, Gaither MR, Hobbs JA, Lozano-Cortes DF, Myers RF, Paulay G, Rocha LA, Toonen RJ, Westneat MW, Berumen ML (2016a) On the origin of endemic species in the Red Sea. J Biogeogr 43:13–30
DiBattista JD, Roberts MB, Bouwmeester J, Bowen BW, Coker DJ, Lozano-Cortes DF, Choat JH, Gaither MR, Hobbs JA, Khalil MT, Kochzius M, Myers RF, Paulay G, Robitzch VSN, Saenz-Agudelo P, Salas E, Sinclair-Taylor TH, Toonen RJ, Westneat MW, Williams ST, Berumen ML (2016b) A review of contemporary patterns of endemism for shallow water reef fauna in the Red Sea. J Biogeogr 43:423–439
Dixon GB, Davies SW, Aglyamova GA, Meyer E, Bay LK, Matz MV (2015) Genomic determinants of coral heat tolerance across latitudes. Science 348:1460–1462
Etter PD (2011) SNP discovery and genotyping for evolutionary genetics using RAD sequencing. Methods Mol Biol 772:157–178
Foll M, Gaggiotti O (2008) A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180:977–993
Gavrilets S (2004) Genetic theories of allopatric and parapatric speciation. In: Dieckmann U, Doebeli M, Metz J, Tautz D (eds) Adaptive speciation. Cambridge University Press, Cambridge, pp 112–139
Giles EC, Saenz-Agudelo P, Hussey NE, Ravasi T, Berumen ML (2015) Exploring seascape genetics and kinship in the reef sponge Stylissa carteri in the Red Sea. Ecol Evol 5:2487–2502
Graham EM, Baird AH, Connolly SR (2008) Survival dynamics of scleractinian coral larvae and implications for dispersal. Coral Reefs 27:529–539
Hoey AS, Feary DA, Burt JA, Vaughan G, Pratchett MS, Berumen ML (2016) Regional variation in the structure and function of parrotfishes on Arabian reefs. Mar Pollut Bull 105:524–531
Howells EJ, Abrego D, Meyer E, Kirk NL, Burt JA (2016) Host adaptation and unexpected symbiont partners enable reef-building corals to tolerate extreme temperatures. Glob Chang Biol 22:2702–2714
Howells EJ, Bauman AG, Vaughan GO, Hume BC, Voolstra CR, Burt JA (2020) Corals in the hottest reefs in the world exhibit symbiont fidelity not flexibility. Mol Ecol 29:899–911
Hume BC, D’Angelo C, Burt JA, Wiedenmann J (2018) Fine-scale biogeographical boundary delineation and sub-population resolution in the Symbiodinium thermophilum coral symbiont group from the Persian/Arabian Gulf and Gulf of Oman. Front Mar Sci 5:138
Kampf J, Sadrinasab M (2006) The circulation of the Persian Gulf: a numerical study. Ocean Sci 2:27–41
Kimura M (1983) The Neutral Theory of Molecular Evolution. Cambridge University Press, Cambrige
Kinlan B, Gaines S (2003) Propagule dispersal in marine and terrestrial environments: a community perspective. Ecology 84:2007–2020
Lambeck K (1996) Shoreline reconstructions for the Persian Gulf since the last glacial maximum. Earth Planet Sci Lett 142:43–57
Leadley P, Pereira HM, Alkemade R, Fernandez-Manjarres JF, Proença V, Scharlemann JPW, Walpole MJ (2010) Biodiversity scenarios: projections of 21st century change in biodiversity, and associated ecosystem services: a technical report for the global biodiversity outlook 3 (No. 50). UNEP/Earthprint
Liew YJ, Howells EJ, Wang X, Michell CT, Burt JA, Idaghdour Y, Aranda M (2020) Intergenerational epigenetic inheritance in reef-building corals. Nat Clim Change 10:254–259
Lischer HE, Excoffier L (2012) PGDSpider: an automated data conversion tool for connecting population genetics and genomics programs. Bioinformatics 28:298–299
Lokier SW, Bateman MD, Larkin NR, Rye P, Stewarte JR (2015) Late Quaternary sea-level changes of the Persian Gulf. Quat Res 84:69–81
Meirmans PG, van Tienderen PH (2004) Genotype and Genodive: Two Programs for the Analysis of Genetic Diversity of Asexual Organisms. Mol Ecol Notes 4:792–794
Munday PL, Leis JM, Lough JM, Paris CB, Kingsford MJ, Berumen ML, Lambrechts J (2009) Climate change and coral reef connectivity. Coral Reefs 28:379–395
Nanninga GB, Saenz-Agudelo P, Manica A, Berumen ML (2014) Environmental gradients predict the genetic population structure of a coral reef fish in the Red Sea. Mol Ecol 23:591–602
Nei M (1987) Molecular Evolutionary Genetics. Columbia University Press, New York
Osman EO, Smith DJ, Ziegler M, Kürten B, Conrad C, El-Haddad KM, Voolstra C, Suggett DJ (2018) Thermal refugia against coral bleaching throughout the northern Red Sea. Global Change Biol 24:e474–e484
Palumbi SR, Barshis DJ, Traylor-Knowles N, Bay RA (2014) Mechanisms of reef coral resistance to future climate change. Science 344:895–898
Paparella F, Xu C, Vaughan GO, Burt JA (2019) Coral Bleaching in the Persian/Arabian Gulf Is Modulated by Summer Winds. Front Mar Sci 6:205
Paris JR, Stevens JR, Catchen JM (2017) Lost in parameter space: a road map for STACKS. Methods Ecol Evol 8:1360–1373
Pratchett MS, Baird AH, Bauman AG, Burt JA (2017) Abundance and composition of juvenile corals reveals divergent trajectories for coral assemblages across the United Arab Emirates. Mar Pollut Bull 114:1031–1035
Priest MA, DiBattista JD, McIlwain JL, Taylor BM, Hussey NE, Berumen ML (2016) A bridge too far: dispersal barriers and cryptic speciation in an Arabian Peninsula grouper (Cephalopholis hemistiktos). J Biogeogr 43:820–832
Riegl B, Purkis SJ (2015) Coral population dynamics across consecutive mass mortality events. Global Change Biol 21:3995–4005
Riegl BM, Benzoni F, Samimi-Namin K, Sheppard C (2012) The Hermatypic Scleractinian (Hard) Coral Fauna of the Gulf. In: Riegl BM, Purkis SJ (eds) Coral Reefs of the Gulf: Adaptation to Climatic Extremes. Springer, Netherlands, Dordrecht, pp 187–224
Riegl B, Johnston M, Purkis S, Howells E, Burt J, Steiner SCC, Sheppard CRC, Bauman A (2018) Population collapse dynamics in Acropora downingi, an Arabian/Persian Gulf ecosystem-engineering coral, linked to rising temperature. Glob Change Biol 24:2447–2462
Rochette NC, Catchen JM (2017) Deriving genotypes from RAD-seq short-read data using Stacks. Nat Protoc 12:2640–2659
Sarnthein M (1972) Sediments and history of the Postglacial transgression in the Persian Gulf and northwest Gulf of Oman. Mar Geol 12:245–266
Shanks A, Grantham B, Carr M (2003) Propagule dispersal distance and the size and spacing of marine reserves. Ecol Appl 13:S159–S169
Sheppard C (1993) Physical Environment of the Gulf Relevant to Marine Pollution: An Overview. Mar Pollut Bull 27:3–8
Sheppard C (2016) Coral reefs in the Gulf are mostly dead now, but can we do anything about it? Mar Pollut Bull 105:593–598
Sheppard CRC, Sheppard ALS (1991) Corals and coral communities of Arabia. Faun Saudi Arab 12:3–170
Sheppard CRC, Price ARG, Roberts CM (1992) Marine Ecology of the Arabian Region: Patterns and Processes in Extreme Tropical Environments. Academic Press, London
Smith EG, Hume BC, Delaney P, Wiedenmann J, Burt JA (2017) Genetic structure of coral-Symbiodinium symbioses on the world’s warmest reefs. PloS one 12:e0180169
Spaet JL, Jabado RW, Henderson AC, Moore AB, Berumen ML (2015) Population genetics of four heavily exploited shark species around the Arabian Peninsula. Ecol Evol 5:2317–2332
Swift SA, Bower AS (2003) Formation and circulation of dense water in the Persian/Arabian Gulf. J Geophys Res 108(C1):3004
Teller JT, Glennie KW, Lancaster N, Singhvi AK (2000) Calcareous dunes of the United Arab Emirates and Noah’s Flood: the postglacial reflooding of the Persian (Arabian) Gulf. Quat Res 68:297–308
Torda G, Donelson JM, Aranda M, Barshis DJ, Bay L, Berumen ML, Bourne DG, Cantin N, Foret S, Matz M, Miller DJ, Moya A, Putnam HM, Ravasi T, van Oppen MJH, Thurber RV, Vidal-Dupiol J, Voolstra CR, Watson SA, Whitelaw E, Willis BL, Munday PL (2017) Rapid adaptive responses to climate change in corals. Nat Clim Change 7:627–636
Torquato F, Møller PR (2020) Physical-biological interactions underlying the phylogeographic patterns of coral-dependent fishes around the Arabian Peninsula. Biorxiv. https://doi.org/10.1101/2020.01.27.921411
Torquato FO, Range P, Ben-Hamadou R, Sigsgaard EE, Thomsen PF, Riera R, Berumen ML, Burt JA, Feary DA, Marshell A, D’Agostino D, DiBattista JD, Møller PR (2019) Consequences of marine barriers on genetic diversity of the coral-specialist yellowbar angelfish from the western Indian Ocean. Ecol Evol 9:11215–11226
van Herwerden L, McIlwain J, Al-Oufi H, Al-Amry W, Reyes A (2006) Development and application of microsatellite markers for Scomberomorus commerson (Perciformes; Teleostei) to a population genetic study of Arabian Peninsula stocks. Fish Res 79:258–266
Vaughan GO, Burt JA (2016) The changing dynamics of coral reef science in Arabia. Mar Pollut Bull 105:441–458
Vaughan GO, Al-Mansoori N, Burt JA (2019) The Arabian Gulf. In: Sheppard C (ed) World seas: An environmental evaluation. Academic Press, Massachuetts, pp 1–23
Wallace CC (1999) Staghorn corals of the world: a revision of the coral genus Acropora (Scleractinia; Astrocoeniina; Acroporidae) worldwide, with emphasis on morphology, phylogeny and biogeography. CSIRO Publishing, Collingwood, Australia
Wallace CC, Done BJ, Muir PR (2012) Revision and Catalogue of Worldwide Staghorn Corals Acropora and Isopora (Scleractina: Acroporidae) in the Museum of Tropical Queensland. Queensland Museum, Brisbane, Australia
Wilson S, Fatemi SMR, Shokri MR, Claereboudt M (2002) Status of coral reefs of the Persian/Arabian Gulf and Arabian Sea region. In: Sheppard C (ed) World seas: An environmental evaluation. Academic Press, Massachuetts, pp 53–62
Acknowledgements
We are thankful to Cécile Richard for supporting the collection around Halul. We are also thankful for the valuable comments provided by the two anonymous reviewers. We appreciate the permits to collect biological material for scientific purposes for the coral samples in Qatar, from the Ministry of Municipality and Environment (Permit Numbers: 73219/2016 and 213126/2017) and in Oman from the Director General of Nature Conservation, Ministry of Environment and Climate Affairs (Permit Number: 6210/10/48).
Funding
Open access funding provided by the Qatar National Library. This work was conducted within the framework of the NPRP projects ‘Connectivity, diversity and genetics between offshore natural coral reefs and oil platforms—NPRP7-1129-1-201’ and ‘Integrated assessment of Qatari coral ecosystems: Toward an Ecosystem-based Approach for management—NPRP8-952-1-186’ both funded by the Qatar National Research Fund (a member of The Qatar Foundation). FT, first author, was supported by a CNPq/Brazil fellowship through the program Science without Borders (Proc. 232875/2014-6). JB was supported by funding for the Water Research Center at NYUAD, Project CG007.
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Topic Editor Francesca Benzoni
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Torquato, F., Bouwmeester, J., Range, P. et al. Population genetic structure of a major reef-building coral species Acropora downingi in northeastern Arabian Peninsula. Coral Reefs 41, 743–752 (2022). https://doi.org/10.1007/s00338-021-02158-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-021-02158-y