Abstract
Polyp bail-out is a stress response exhibited by some pocilloporid corals, with mechanisms and consequences distinct from those of bleaching. Although induction of polyp bail-out has been demonstrated in the laboratory, molecular mechanisms underlying this response have rarely been discussed. We conducted genetic analyses of Pocillopora acuta during initiation of hyperosmosis-induced polyp bail-out, using both transcriptomic and qPCR techniques. Beyond upregulation of apoptosis and proteolysis, corals showed significant activation of tumor necrosis factor and fibroblast growth factor (FGF) signaling pathways during induction of polyp bail-out. In our qPCR analysis, a common upregulation profile, peaking at 43.0‰ salinity, was found in the FAS and CASP8 genes, whereas a different profile, showing significant upregulation up to 45.0‰, was displayed by matrix metalloproteinases and genes in the FGF signaling pathway. These results suggest parallel involvement of an extrinsic apoptotic signaling pathway and FGF-mediated extracellular matrix degradation in polyp bail-out. Furthermore, in the XIAP, JNK, and NFKB1 genes, we detected a third expression profile showing linear upregulation that becomes maximal at the endpoint salinity level of the experiment (46.0‰), indicating activation of anti-apoptotic and cell survival signals during polyp bail-out. Our results provide new insights into signaling pathways responsible for polyp bail-out and suggest the feasibility of inducing bail-out by specifically triggering these pathways without exerting lethal stresses on the corals, which in turn will facilitate acquisition of viable polyps for possible use in coral reef restoration.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Corals in the family Pocilloporidae (Cnidaria: Anthozoa: Scleractinia) are among the major tropical reef-building corals in the Indo-Pacific Ocean (Pinzon and LaJeunesse 2011). As pocilloporid corals generally inhabit shallow water, which renders them susceptible to thermal and osmotic fluctuations (Schmidt-Roach et al. 2014; Poquita-Du et al. 2019), studies of stress responses in pocilloporids are of great importance to understand anthropogenic impacts on coral reef ecosystems. Coral bleaching, which is the systematic dissociation of endosymbiotic dinoflagellates from their coral hosts, is a general response of stony corals to a vast number of environmental stresses (Lesser et al. 1990; Jones and Hoegh-Guldberg 1999; Jones 2005; Ainsworth et al. 2008). As most stony corals rely on photosynthesis from symbiotic algae to meet their carbon requirements, coral bleaching is responsible for the high mortality rate among tropical corals (Hoegh-Guldberg 1999; Carpenter et al. 2008).
In recent decades, a novel stress response called “polyp bail-out” has been increasingly reported in pocilloporid corals (Sammarco 1982; Domart-Coulon et al. 2004; Kvitt et al. 2015; Shapiro et al. 2016; Fordyce et al. 2017; Wecker et al. 2018). Unlike coral bleaching, polyp bail-out is characterized by dissociation of coral colonies via coenosarc degradation and detachment of zooxanthellate polyps from the calcareous skeletons (Sammarco 1982). In natural environments, polyp bail-out has been reported in the Great Barrier Reef and reefs along the Pacific coast of Costa Rica, although causes are uncertain (Sammarco 1982; Wild et al. 2014). Under laboratory conditions, induction of polyp bail-out has been demonstrated with different treatments, including calcium deprivation, acidification, thermal stress, hyperosmosis, starvation, and insecticides (Domart-Coulon et al. 2004; Kvitt et al. 2015; Shapiro et al. 2016; Fordyce et al. 2017; Serrano et al. 2018; Wecker et al. 2018). As detached polyps can be maintained in the laboratory for weeks to months (Capel et al. 2014; Shapiro et al. 2016; Serrano et al. 2018), these polyps offer subject material for in situ studies of coral cellular biology and of symbiotic relationships between coral animals and symbiotic algae. Moreover, since detached polyps are able to resettle and resume skeletogenesis after stress abates, polyp bail-out can be considered an asexual reproductive method in stony corals and may provide an alternative approach to mass production of coral colonies for reef restoration (Sammarco 1982; Shapiro et al. 2016; Fordyce et al. 2017).
However, documented success of polyp survival and resettlement after bail-out varies greatly among studies (Sammarco 1982; Shapiro et al. 2016; Fordyce et al. 2017). These variations may be attributable to treatments used in different studies and to the quality of resettling environments, as well as the health of the corals in question (Shapiro et al. 2016). Understanding molecular mechanisms responsible for polyp bail-out is the first step in determining how different stimuli affect the process and consequences of this stress response. Identifying core signaling pathways leading to the response will also help to develop methods to induce the response with minimal impact upon coral health, which in turn will facilitate survival and resettlement of detached polyps. Recently, genomes and transcriptomes of some pocilloporid corals have been published (Traylor-Knowles et al. 2011; Voolstra et al. 2017; Cunning et al. 2018), enabling a more thorough understanding of this response from a molecular perspective. A recent transcriptomic study of Pocillopora damicornis during polyp bail-out demonstrated overexpression of many caspase-encoding genes in concert with coenosarc degradation (Wecker et al. 2018), supporting the hypothetical link between polyp bail-out and tissue-specific apoptosis revealed previously in a protein activity assay (Kvitt et al. 2015). In addition, Wecker et al. (2018) proposed that proteolytic enzymes, such as cathepsins, trigger degradation of the extracellular matrix (ECM) between coral polyps and the calcareous skeletons, resulting in detachment of polyps. However, our understanding of molecular mechanisms in polyp bail-out, particularly signaling pathways activating apoptotic and proteolytic responses, is still in its infancy.
In the present study, we employed hyperosmotic stress to induce bail-out in P. acuta, a species closely related to P. damicornis, according to recent phylogenetic classifications (Schmidt-Roach et al. 2014; Johnston et al. 2017). Based on both transcriptomic and qPCR techniques, we sought to identify the signaling pathways associated with polyp bail-out.
Materials and methods
Coral collection and maintenance
During 2018–2019, we purchased nine colonies of Pocillopora acuta from the Onna Village Fisheries Cooperative in Okinawa, Japan. Coral colonies were transferred to the OIST Marine Science Section at Seragaki, where they were kept for over 6 months in a 3000-L outdoor tank supplied with flowing, sand-filtered natural seawater, before the commencement of polyp bail-out experiments.
Polyp bail-out induction and RNA sampling for transcriptomic analysis (Experiment I)
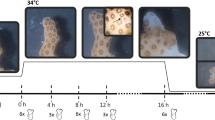
In order to examine molecular mechanisms underlying polyp bail-out, we induced the bail-out response in P. acuta using a hyperosmotic treatment modified from Shapiro et al. (2016). One day prior to the experiment, eight coral fragments (0.5–1 cm in length) from a mother colony were clipped off and randomly placed in two 5-L indoor experimental tanks (four in the treatment group and four in the control group). For both experimental tanks, light intensity was provided at 300 μmol photons m−2 s−1 (measured with an Apogee MQ-210 underwater quantum meter) with a 12-h day–night cycle. Artificial seawater (Kaisuimaren, Japan) at 35‰ salinity was prepared in both tanks, and water temperature was allowed to fluctuate daily in the range of 24–26 °C. Evaporation-driven hyperosmotic treatment, as reported in Shapiro et al. (2016), not only increases seawater salinity, but concentrates coral metabolic waste and microorganisms in seawater, which presumably constitute another stress on corals. Therefore, in the present study, we induced hyperosmotic stress by addition of hypersaline seawater. For the treatment group, high-salinity artificial seawater (48‰) was pumped into the experimental tank at a constant rate of 4 mL min−1 using a peristaltic pump, while for the control group, ambient salinity (35‰) was maintained. At 0 h, 12 h, and 24 h of the experiment, seawater salinity and temperature were monitored with a ProfiLux 4 aquarium controller (GHL, Germany) and coral morphological changes were recorded using a stereo microscope (Leica, Germany). To examine genetic responses during initiation of polyp bail-out, one coral fragment was sampled at each of the time points from both treatment (samples labeled as salinity levels at the sampling points: T35.0, T43.0, and T46.0) and control groups (C35.0, C35.4, and C35.8) for subsequent Illumina RNA sequencing (Fig. 1). Since partial polyp detachment (an indicator of onset of polyp bail-out) was observed in the treatment group at 46.0‰ salinity (24 h) and bailed-out polyps could be easily removed from the skeleton by gently shaking the coral fragment in water, we collected both undetached coral tissues (whole fragment including undetached polyps, remaining coenosarc, and underlying skeleton; T46.0-U) and detached polyps (10–15 completely detached polyps, collected with a large-bore pipette; T46.0-D) separately. Coral samples were immediately preserved in 1 mL TRIZOL reagent (Thermo Fisher Scientific, USA) after collection and were stored at − 20 °C until RNA extraction. To avoid artifacts from microscopic observation, a spare coral fragment in both treatment and control groups was used for morphological observation throughout the experiment and was not included in subsequent genetic analysis. From the spare coral fragment in the treatment group, we transferred detached polyps to the control tank immediately after their detachment. These polyps were later examined for viability via a stereo microscope (Leica, Germany). The same experiment was repeated three times on different P. acuta colonies, with at least 1 month between experiments (N = 3).
Polyp bail-out in Pocillopora acuta under hyperosmotic stress. Seawater salinity changes in treatment and control groups during the experiment are presented in the central plot with black and gray lines, respectively. Coenosarc thinning/degradation (arrow) commenced in the treatment group when salinity reached 43.0‰ (12 h). Polyp detachment (yellow circle) was observed in the treatment group at 46.0‰ salinity (24 h), yielding solitary, morphologically intact polyps (upper right box of treatment group at 46.0‰). Coral morphological changes were recorded on a spare coral fragment in both treatment and control groups at each sampling point using a stereo microscope
RNA extraction, Illumina RNA sequencing, and transcriptome assembly
For all tissue samples collected in this study, soft tissues were removed from the skeleton (if applicable) and homogenized in the TRIZOL reagent using a beadblaster tissue homogenizer (2500 rpm for 2 min) before RNA extraction. Total RNA was extracted following the TRIZOL RNA extraction protocol (Chomczynski 1993), and RNA quality was checked with a Bioanalyzer 2100 (Agilent, Japan). For Experiment I, RNA samples with RNA integrity number (RIN) ≥ 7 were sent to the DNA Sequencing Section at the Okinawa Institute of Science and Technology (OIST) for library construction (strand-specific and polyA-RNA purified; number of libraries = 7 experimental conditions × 3 biological replicates) and Illumina HiSeq 4000 150 × 150-bp paired-end RNA sequencing. Raw reads from Illumina RNA sequencing were trimmed with Trimmomatic v0.36 (Bolger et al. 2014) to remove adapters and low-quality sequences and were checked for quality using FastQC (Andrews 2010). To remove RNA reads from Symbiodinium and other coral-associated microbiomes, we first mapped trimmed RNA reads (~ 877 million read pairs from all sequenced libraries) to a reference Pocillopora damicornis genome assembly (GenBank assembly accession: GCA_003704095.1) (Cunning et al. 2018) using Tophat v2.1.1 (Kim et al. 2013). Successfully aligned RNA reads (~ 168 million read pairs; successful alignment rate: 19.1%) were then subjected to de novo assembly using Trinity v2.8.4 (Grabherr et al. 2011). RNA read quantification was conducted using RSEM v1.3.2 (Li and Dewey 2011), and reconstructed transcripts shorter than 200 nt or with low coverage (< 5 transcripts per million) were discarded. BUSCO analysis was performed using the metazoa_odb9 dataset to examine completeness of the transcriptome assembly (Simão et al. 2015).
Functional annotation, differential gene expression, and gene ontology enrichment analyses
We performed functional annotation of the transcriptome assembly by searching the SwissProt eukaryotic protein database (downloaded on July 3, 2019) with BLASTX, using a threshold of E = 10−5. Principle component analysis (PCA) and a preliminary differential gene expression (DE) analysis of annotated transcripts showed that RNA-Seq libraries could be clearly classified based on salinity levels, despite a certain level of variation between biological replicates (Fig. 2). We therefore clustered RNA-Seq libraries into three groups, labeled with corresponding salinity levels (S35: T35.0, C35.0, C35.4, C35.8; S43: T43.0; S46: T46.0-U, T46.0-D). A secondary DE analysis was then conducted on annotated transcripts for pairwise comparisons between the three groups (expressed as S35/S43, S35/S46, and S35/S43), designed to cancel out individual variation and to identify transcriptomic responses induced specifically by hyperosmotic treatment. Both preliminary and secondary DE analyses were performed using edgeR (Robinson et al. 2010) with the criteria of > twofold absolute change and > 5 CPM (counts per million) for at least 3 libraries in a given pairwise comparison. To identify cellular processes and signaling pathways responsible for polyp bail-out, gene ontology (GO) enrichment analysis was performed for differentially expressed genes (DEG) using DAVID bioinformatics resources v6.8 (Huang et al. 2008, 2009).
Principal component analysis and gene expression heatmap. Principal component analysis was conducted based on 17,908 functionally annotated transcripts. PC1 is coincident with salinity gradient (highlighted with a red-to-white gradient) and clusters RNA-Seq libraries into three groups based on salinity levels (S35: C35.0, C35.4, C35.8, T35.0; S43: T43.0; S46: T46.0-U, T46.0-D) (a), within which variations between biological replicates (Rep) are reflected by PC2 and PC3 (b). A gene expression heatmap was plotted based on 4954 differentially expressed genes and shows the same clustering as in the principal component analysis (c). RNA-Seq libraries are labeled: T35.0: treatment group at 35.0‰ (0 h); T43.0: treatment group at 43.0‰ (12 h); T46.0-U: undetached coral tissues in treatment group at 46.0‰ (24 h); T46.0-D: detached polyps in treatment group at 46.0‰ (24 h); C35.0: control group at 35.0‰ (0 h); C35.4: control group at 35.4‰ (12 h); C35.8: control group at 35.8‰ (24 h)
Polyp bail-out induction for real-time quantitative PCR analysis (Experiments II and III)
To validate polyp bail-out and genetic responses induced by hyperosmosis, in 2019–2020 we repeated the polyp bail-out experiment under: (1) the same culturing conditions as Experiment I (artificial seawater at 35‰; 300 μmol photons m−2 s−1; Experiment II) and (2) different culturing conditions (filtered seawater at ~ 35‰; 150 μmol photons m−2 s−1; Experiment III). In each experiment, 36 fragments from three P. acuta colonies (N = 3; 12 fragments/colony) were randomly placed in two 5-L tanks 1 day prior to the experiment (6 in treatment group and 6 in control group for each colony). For better temporal resolution of genetic analysis, one fragment/colony was sampled from both treatment and control groups every 6 h during the experiment (Fig. 1), at which salinity levels (treatment group/control group) were 35.0‰/35.0‰ (0 h), 40.0‰/35.2‰ (6 h), 43.0‰/35.4‰ (12 h), 45.0‰/35.6‰ (18 h), and 46.0‰/35.8‰ (24 h). As in Experiment I, seawater salinity, temperature, and coral morphological changes (in the spare fragments) were recorded at each sampling point. Since the transcriptomes of undetached coral tissues (T46.0-U) and detached polyps (T46.0-D) collected in Experiment I showed no obvious differences in our preliminary analyses (Fig. 2 and Table S1), in Experiments II and III we collected undetached coral tissues and detached polyps together as one sample (treatment group; 46.0‰). Collected coral samples were preserved in TRIZOL reagent as in Experiment I and were subsequently subjected to real-time quantitative PCR (qPCR) analysis.
cDNA synthesis and qPCR assay
For samples from Experiments II and III (N = 3 biological replicates × 5 sampling points × 2 experimental conditions (treatment vs. control) for each experiment), total RNA was extracted as described above and cDNA synthesis was conducted using the SuperScript IV VILO Master Mix (Invitrogen, USA). Based on our transcriptomic data, we designed a qPCR assay to examine expression profiles of 10 stress genes during polyp bail-out induction (Table 1). Genes selected belong to the extrinsic apoptotic signaling pathway (FAS and CASP8), the fibroblast growth factor signaling pathway (FGF2 and FGFR2), the Ras signaling pathway (RHO, a Ras-like GTP-binding protein), or participate in proteolysis (MMP19 and MMP24), or in anti-apoptotic/survival signaling (JNK, NFKB1, and XIAP). A β-tubulin gene (TUBB) was included as an internal control gene for the qPCR analysis. For genes identified in multiple transcripts, the transcript with the highest expression level was selected as the representative in the qPCR assay, based on the assumption that such transcripts had the greatest biological significance. For each qPCR reaction, 200 μM of each primer and 1 μL of synthesized cDNA (concentration undetermined) were mixed with iQ SYBR Green Supermix (Bio-Rad, USA) to a total volume of 10 μL. Specificity of primer pairs was confirmed by a melting curve analysis (MCA) and qPCR efficiencies were tested in 10x-serial dilution with at least four concentration points (N = 3 at each concentration). All qPCR reactions were performed on a StepOnePlus Real-Time PCR System (Thermo Fisher Scientific, USA) under the following conditions: an initiation step at 95 °C for 3 min followed by 40 cycles of PCR amplification at 95 °C for 15 s and 60 °C for 30 s. For each gene in the qPCR assay, the mean cycle threshold (CT) value from two technical replicates of a given sample was employed to calculate the relative gene expression (∆CT = CTcontrol gene − CTtarget gene). As qPCR results from both Experiments II and III showed the same trend of expression changes with only slight variation in magnitude (Fig. S1 and Table S2), data from the two experiments were pooled to increase statistical power. The ∆CT values of treatment group were normalized to those of the corresponding control group (on an average of six replicates) and to the initial conditions (35.0‰) to determine expression changes during the experiments (∆∆∆CT = ∆∆CTgiven salinity level − ∆∆CTinitial condition; ∆∆CT = ∆CTtreatment group − ∆CTcontrol group). Data are presented as means ± SDs (standard deviations) for each gene, and statistical significance between time points was tested using Welch’s ANOVA with a Games-Howell post hoc test.
Results
Polyp bail-out induction
To induce the polyp bail-out response in Pocillopora acuta, we established a hyperosmotic environment by gradual addition of high-salinity seawater (48‰). Disregarding differences in lighting and water conditions among the three experiments in this study, the same salinity-change profiles and morphological changes were observed (Fig. 1). In the treatment group (Fig. 1, upper panels), tentacle retraction was observed when salinity increased from 40.0 to 43.0‰ (6–12 h). Coenosarc tissues started to thin/degrade after salinity reached 43.0‰ (12 h) and became more perceptible at 45.0‰ salinity (18 h). At 46.0‰ salinity (24 h), complete coenosarc degradation was observed at some part of the coral fragments and about 30–50% of the polyps were found detached from the skeletons, apparently intact. To examine viability of polyps after the bail-out response, in each experiment we transferred some detached polyps to the control tank immediately after their detachment. In all these polyps, viability was evidenced by rotatory movement after few hours of rescue, which could be maintained for over 24 h (supplementary video). For control groups in all the three experiments, salinity changes during the experiment period were < 1‰ and no sign of polyp bail-out or other stress response was observed during the experiments (Fig. 1, lower panels).
Transcriptome assembly and differential gene expression analysis
After filtering RNA reads from coral-associated microbiomes and removing transcripts of low coverage, 73,903 transcripts were reconstructed (N50 = 1278 nt; E90N50 = 1746 nt; GenBank assembly accession: GIDI00000000), with 84% transcriptomic completeness (fragmented, 9.7%; missing, 6.2%) identified by the BUSCO analysis. From the transcriptome assembly, 17,908 transcripts were functionally annotated as eukaryotic genes. Differential gene expression (DE) analysis identified 6827 differentially expressed genes (DEG) among the three pairwise comparisons (S35/S43, S35/S46, and S43/S46; Table 2), including 11 caspase-like genes and 22 matrix metalloproteinase (MMP)-like genes (Table S3). A Fas receptor (FAS)-like gene (Pacuta_4317_c14_g1_i1) was upregulated in our transcriptomic data, but was statistically insignificant in the DE analysis, due to large variation between biological replicates. A Fas ligand (FASL)-like gene with low coverage was found in the raw transcriptome assembly (Fig. S2) and was removed in the final version of the transcriptome assembly.
In the comparison S35/S43, 3473 DEGs were identified (Table 2), with 1951 showing upregulation (S43 > S35) and 1522 showing downregulation (S43 < S35). In S35/S46, 6483 DEGs were identified (Table 2), with 2874 and 3609 showing upregulation (S46 > S35) and downregulation (S46 < S35), respectively, while in the comparison S43/S46, 99 upregulated (S46 > S43) and 215 downregulated (S46 < S43) DEGs were identified (Table 2). Expression heatmap of the 6827 DEGs further showed three expression profiles that can roughly be characterized as mild upregulation (cluster 1; 2961 DEGs), downregulation (cluster 2; 3788 DEGs), and huge upregulation (cluster 3; 78 DEGs) (Fig. 3).
Differential gene expression analysis for comparisons among S35, S43, and S46. A gene expression heatmap groups 6827 differentially expressed genes (DEG) into three clusters: cluster 1—mild upregulation along the salinity gradient; cluster 2—mild downregulation along the salinity gradient; and cluster 3—huge upregulation along the salinity gradient. Numbers of caspase-like (CASP) and matrix metalloproteinase-like (MMP) genes in each cluster are indicated
Gene ontology enrichment analysis
To identify molecular mechanisms involved in the polyp bail-out response, gene ontology (GO) enrichment analysis was conducted for DEGs identified in the DE analysis. As most enriched GO terms in our results are likely involved in general responses of corals to osmotic fluctuations, the discussion of which is beyond the scope of this study, we paid specific attention to biological processes and signaling pathways related to apoptosis, proteolysis, and cell survival, which have been proposed as participating directly in polyp bail-out (Kvitt et al. 2015; Wecker et al. 2018). Among upregulated DEGs (clusters 1 and 3), GO categories such as protein ubiquitination, tumor necrosis factor (TNF)-mediated signaling pathway, positive regulation of I-kappaB kinase/NF-kappaB signaling, apoptotic process, Ras protein signal transduction, fibroblast growth factor (FGF) receptor signaling pathway, extrinsic apoptotic signaling pathway, and proteolysis were significantly enriched (p < 0.05; Table 3). Among downregulated DEGs (cluster 2), significant enrichment was identified in several GO categories related to cellular metabolism and cell cycle, but no specific signaling pathway or apoptosis/proteolysis-related GO category showed significant enrichment.
Quantitative PCR assay
To achieve finer temporal resolution of genetic analysis and to validate genetic responses identified in our transcriptomic data, we conducted two additional polyp bail-out experiments (Experiments II and III) and collected RNA samples at 6-h intervals for quantitative PCR (qPCR) analysis. For the 10 stress genes in our qPCR assay, gene expression changes showed trends concordant with those identified in the transcriptomic analysis and three expression profiles were consistently identified in both Experiments II and III, despite variations in lighting and water conditions (Fig. S1 and Table S2). For the CASP8 and FAS genes, significant upregulation was identified when salinity increased from 35.0 to 43.0‰ (3.3- and 10.8-fold changes, respectively; Table S2), after which expression remained statistically unchanged (Fig. 4, Profile I). A second expression profile was exhibited by the FGF2, FGFR2, RHO, and the two MMPs genes. For these five genes, little upregulation (related to the magnitudes along the whole experiments) was identified at 35.0–40.0‰ salinity (1.6–11-fold changes; Table S2), but was followed by remarkable upregulation from 40.0 to 45.0‰ (20–1167-fold changes; Table S2), at which salinity upregulation reached its peak (Fig. 4, Profile II). In contrast, for the NFKB1, JNK, and XIAP genes, linear upregulation was detected starting from different salinity levels (35.0–43.0‰) and peaked at the endpoint salinity (46.0‰) of the experiments (Fig. 4, Profile III).
Gene expression profiles of 10 stress genes during polyp bail-out induction. The FAS and CASP8 genes show significant upregulation during salinity increases from 35.0 to 43.0‰ and remain statistically stable at salinity levels of 43.0–46.0‰ (Profile I). The FGFR2, FGF2, RHO, MMP19, and MMP24 genes display a common expression profile that shows little upregulation from 35.0 to 40.0‰, followed by remarkable upregulation from 40.0 to 45.0‰, after which expression stabilizes (Profile II). For the JNK, NFKB1, and XIAP genes, linear upregulation was observed peaking at 46.0‰ salinity (Profile III). Data are labeled as the salinity levels of treatment group at the sampling points. Statistical analysis was conducted using Welch’s ANOVA with a Games-Howell post hoc test. Groups with significant differences (p < 0.05) are indicated. Data are log2-transformed and are expressed as means ± SDs (standard deviations)
Discussion
Hyperosmosis induces polyp bail-out in pocilloporid corals
In three pocilloporid corals, i.e., Pocillopora damicornis, Seriatopora hystrix, and Stylophora pistillata, Shapiro et al. (2016) first showed that polyp bail-out can be induced by evaporation-driven hyperosmotic stress. In the present study, we applied hyperosmotic stress to P. acuta by addition of hypersaline artificial seawater and observed polyp bail-out when seawater salinity reached 46.0‰ after 24 h. Additive salinity increases differ from evaporation-based salinity increases in that the rate of salinity change decreases during the treatment period. This slowdown of stress elevation was reflected in our transcriptomic data, which showed 3473 DEGs in the early stage of experiments (35.0–43.0‰) but only 314 DEGs in the later stage (43.0–46.0‰). Despite this difference in the salinity-change profiles between the present study and Shapiro et al. (2016), polyp bail-out was observed in both studies with the same process and at relatively similar stress levels (11–15‰ higher than ambient conditions). This consistency implies that polyp bail-out may be a common response to extreme hyperosmosis in pocilloporid corals from different habitats. Since detached polyps are viable after hyperosmosis-induced polyp bail-out and are able to resettle once stress abates (Shapiro et al. 2016; Liu et al. 2020), this stress response may represent a mechanism for coral polyps to escape and survive temporary lethal stresses (Sammarco 1982). Additional studies, however, are needed to further illuminate the ecological significance of this stress response.
TNF signaling leads to apoptosis and coenosarc degradation
The extrinsic apoptotic signaling pathway, comprising signals mediated by the TNF receptor family and a caspase cascade initiated by caspase-8, is a highly conserved pathway of programmed cell death among animals (Quistad et al. 2014). In the present study, the significant enrichment of GO terms apoptotic process and extrinsic apoptotic signaling pathway, as well as the coincident upregulation of the FAS and CASP8 genes identified in our genetic analyses, supports the hypothesis of apoptosis-mediated polyp bail-out proposed in previous studies (Kvitt et al. 2015; Wecker et al. 2018). Interestingly, in the present study, overexpression of both the CASP8 and FAS genes peaked at 43.0‰ and remained stable afterward, even through the hyperosmotic stress continued to increase (from 43.0 to 46.0‰ salinity). This plateau of gene expression is coincident with coenosarc thinning/degradation in our morphological observations. Our results thus indicate that corals are able to moderate the apoptotic response once it reaches an appropriate level, adding further support to the hypothesis of “polyp-controlled programmed cell death” in the coenosarc-degradation stage in polyp bail-out (Kvitt et al. 2015).
Interestingly, even with significant enrichment of the TNF signaling pathway and upregulation of the FAS gene in our genetic data, no overexpression of TNF was found during polyp bail-out induction in this study. Similar to our finding, in Wecker et al. (2018) TNF exhibited an expression change opposite to that of caspase and TNF receptor genes during the coenosarc-degradation stage. These observations imply that apoptosis in polyp bail-out is probably disentangled from coral TNF signals. Considering the ancient origin of core apoptotic mechanisms and the growing number of reports on inter-kingdom communication between microbes and their eukaryotic hosts (Hughes and Sperandio 2008; Segovia 2008; Pacheco and Sperandio 2009), we hypothesize that the triggering signal of apoptosis in polyp bail-out is derived from microorganisms associated with corals (Fig. 5). Changing of culture environments can substantially alter the composition of coral-associated microbiomes and the chemical signals provided from microbes to the coral animals (Littman et al. 2011; Sharp and Ritchie 2012; Webster et al. 2013). These coral-associated microorganisms therefore may play an intermediate role between environmental stresses and coral polyp bail-out. In fact, participation of microorganisms in polyp bail-out has recently been demonstrated with induction of the bail-out response in P. damicornis via inoculation with a coral pathogen, Vibrio coralliilyticus (Gavish et al. 2018). Nevertheless, since the TNF signaling pathway is believed to have originated with the Metazoa (Wiens and Glenney 2011; Quistad and Traylor-Knowles 2016) and inter-kingdom communication can proceed via non-coding RNA (Leitão and Enguita 2016; Bayer-Santos et al. 2017), a microbe-derived molecule triggering the coral TNF signaling pathway cannot be identified with the methods used in this study, even in the raw dataset with Symbiodinium/microbiome sequences. Further studies are therefore required to verify this cross-talk and to identify the key signaling molecule that activates the extrinsic apoptotic signaling pathway in coral polyp bail-out.
Putative signaling pathways in polyp bail-out. Environmental stress triggers the TNF and FGF signaling pathways, which activate the caspase (CASP) and matrix metalloproteinase (MMP) cascades, respectively. Activation of caspases leads to apoptosis in the coenosarc and to its subsequent degradation, while activation of MMPs, likely through the Ras signaling pathway (RAS), results in extracellular matrix (ECM) degradation and detachment of polyps. In polyp bodies, anti-apoptotic and cell survival signals mediated by genes, such as JNK, NFKB, and XIAP, are activated to suppress the apoptotic response, which in turn promotes polyp survival
FGF signaling induces ECM degradation and polyp detachment
A previous study of P. damicornis suggested participation of ECM degradation in the polyp-detachment step of bail-out response (Wecker et al. 2018). Congruent with that study, we found significant enrichment of proteolysis and remarkable expression changes of several MMP-encoding transcripts during polyp bail-out induction. MMPs are a family of proteases that function in ECM degradation (Birkedal-Hansen et al. 1993; Kojima et al. 2000). Regulation of these MMPs in polyp bail-out is probably linked to the FGF and Ras signaling pathways, based on (1) the concurrent enrichment of GO categories FGF receptor signaling pathway and Ras protein signal transduction in our transcriptomic data, and (2) matched expression profiles between the FGF2, FGFR2, RHO, and the two MMP genes in our qPCR analysis. In many mammalian cells, the FGF-Ras-MMP signaling pathway is involved in cell migration, angiogenesis, and cancer metastasis (Liu et al. 2002; Pintucci et al. 2003; Krejci et al. 2005). Since upon activation, some MMPs also induce release of FGF2 (Whitelock et al. 1996), this feedback circuit between MMPs and FGF signals may explain the remarkable upregulation of FGF2 and MMPs at 40.0–45.0‰ in our qPCR analysis, and the irreversibility of coral polyp bail-out. Accordingly, we hypothesize that in polyp bail-out, FGF signaling is likely relayed by the Ras signaling pathway to activate MMPs that initiate subsequent ECM degradation and polyp detachment (Fig. 5).
Survival mechanisms of polyps during the bail-out response
Based on histological data, Kvitt et al. (2015) demonstrated that the apoptotic response in polyp bail-out occurs mostly in the coenosarc and adjacent tissues, but not in polyps. However, in our transcriptomic data, no significant difference in caspase gene expression was found between coenosarc-containing coral tissues (T46.0-U) and detached solitary polyps (T46.0-D). We therefore hypothesize that tissue-specific apoptosis in polyp bail-out is mediated at the posttranscriptional level. In support of this hypothesis, in our transcriptomic data we identified significant enrichment of GO category protein ubiquitination and upregulation of the XIAP gene, which have both been shown to regulate caspase activity and cell death (Deveraux et al. 1997; Bader and Steller 2009; Chen and Qiu 2013). It has been proposed that an anti-apoptotic response is activated by TNF signals at a later stage of polyp bail-out, based on the coincident upregulation of TNF genes and downregulation of caspases and TNFRs after coenosarc degradation (Wecker et al. 2018). Similarly, in the present study, enrichment of GO terms such as JNK cascade, negative regulation of extrinsic apoptotic signaling pathway via death domain receptors, and positive regulation of I-kappaB kinase/NF-kappaB signaling was identified among upregulated DEGs, indicating activation of anti-apoptotic/cell survival responses during polyp bail-out (Van Antwerp et al. 1996; Roulston et al. 1998; Karin and Lin 2002). These responses, however, were not accompanied by TNF signals, as our transcriptomic analysis showed no TNF overexpression during the experiment, even in detached polyps (T46.0-D). Alternatively, survival and anti-apoptotic signals may be associated with cellular stresses accumulated during polyp bail-out, implied by the significant enrichment of GO category response to oxidative stress and by linear upregulation of survival and anti-apoptotic genes along salinity increases in our qPCR experiments. Accordingly, our results suggest that the tissue-specificity of apoptosis in polyp bail-out may be attributed to spatial variation in anti-apoptotic signals, leading to inhibition of caspase activity in coral polyps and to their subsequent survival (Fig. 5). Identifying the ultimate trigger of these anti-apoptotic/cell survival signals and their functional involvement in polyp bail-out, however, requires further investigation.
New insights distinguish polyp bail-out and coral bleaching
Based on our results, it appears that the TNF receptor-mediated extrinsic apoptotic pathway, FGF-mediated ECM degradation, and anti-apoptotic/survival signals are involved in polyp bail-out (Fig. 5). Bail-out is distinct from coral bleaching, which is generally thought to be mediated via the intrinsic apoptotic pathway triggered by oxidative stress from dinoflagellate symbionts (Weis 2008; Kvitt et al. 2011). The unique activation of FGF-mediated ECM degradation in polyp bail-out further distinguishes it from bleaching with respect to morphological consequences. In polyp bail-out, free-living polyps are generated while in bleaching, azooxanthellate coral polyps remain in situ after stress. As some environmental stresses, such as acidification and thermal stress, are common inducers for both responses, understanding mechanisms by which corals determine which stress response is to be employed, is essential to understand the relative ecological significance of polyp bail-out versus coral bleaching.
Future applications of polyp bail-out
Beside its distinctness from coral bleaching, the discreteness of pathways leading to apoptosis and ECM degradation in bail-out suggests possible independence of coenosarc degradation and polyp detachment. Indeed, separation of these processes has been reported in previous studies using different treatments (Domart-Coulon et al. 2004; Kvitt et al. 2015; Fordyce et al. 2017) and was observed in our preliminary experiments under different levels of hyperosmotic stresses (Fig. S3). Also, since detachment of polyps is mediated by degradation of ECM, we hypothesize that it may be possible to induce polyp resettlement either by stimulating ECM secretion or by providing suitable ECM components to bailed-out polyps. In support of our hypothesis, previous studies have found that fibronectin-like peptide facilitated attachment of solitary polyps to resettlement substrates (Domart-Coulon et al. 2004; Puverel et al. 2005). However, in both Domart-Coulon et al. (2004) and Puverel et al. (2005), coral polyps tended to dissociate into single cells after the resettlement. This cell dissociation was likely caused by calcium-deprivation treatments used to detached polyps in both studies, which might be expected to impact cell–cell adhesion (Hobmayer et al. 2001; Marshall and Clode 2004; Van Roy and Berx 2008). In contrast, hyperosmosis-based polyp bail-out induction offers advantages over Ca-deprivation in maintaining morphological integrity of polyps after their resettlement (Shapiro et al. 2016; Liu et al. 2020). However, the extreme hyperosmotic stress applied during polyp bail-out induction could impair coral health and reduce resettlement success of detached polyps (Shapiro et al. 2016). Our identification of signaling pathways underlying the bail-out response sheds light on development of alternative methods for polyp bail-out induction, such as using ligands or agonists specifically targeting these pathways. By minimizing systematic stress and avoiding undesirable responses in corals, new methods are expected to facilitate acquisition of resettled polyps that are morphologically and physiologically intact. Given that polyp bail-out from a small coral fragment can generate tens to hundreds of individual polyps (Sammarco 1982; Shapiro et al. 2016; Fordyce et al. 2017), this coral propagation method provides potentially significant advantages over the commonly used cutting/fragmentation strategy in regard to the output quantity. Improvement of success rates in resettlement and survival of bailed-out polyps will potentiate their use for mass production of coral colonies for reef restoration. Furthermore, since polyp bail-out may greatly influence local recruitment in a variety of coral species (Fordyce et al. 2017), our findings provide a genetic perspective for future studies to understand how this response shapes coral reef ecosystems in shallow waters.
Data accessibility
Data supporting the findings of this study are available on the NCBI database with the following accession numbers: RNA-Seq raw data: NCBI SRA: SRR10696823-SRR10696829, SRR10708182-SRR10708188, SRR10708225-SRR10708230. Transcriptome assembly: GenBank assembly accession GIDI00000000.
References
Ainsworth T, Hoegh-Guldberg O, Heron S, Skirving W, Leggat W (2008) Early cellular changes are indicators of pre-bleaching thermal stress in the coral host. Journal of Experimental Marine Biology and Ecology 364:63–71
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. Babraham Bioinformatics, Babraham Institute, Cambridge, United Kingdom
Bader M, Steller H (2009) Regulation of cell death by the ubiquitin–proteasome system. Current Opinion in Cell Biology 21:878–884
Bayer-Santos E, Marini MM, da Silveira JF (2017) Non-coding RNAs in host–pathogen interactions: subversion of mammalian cell functions by protozoan parasites. Frontiers in Microbiology 8:474
Birkedal-Hansen H, Moore W, Bodden M, Windsor L, Birkedal-Hansen B, DeCarlo A, Engler J (1993) Matrix metalloproteinases: a review. Critical Reviews in Oral Biology & Medicine 4:197–250
Bolger AM, Lohse M, Usadel B (2014) Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120
Capel KCC, Migotto A, Zilberberg C, Kitahara MV (2014) Another tool towards invasion? Polyp “bail-out” in Tubastraea coccinea. Coral Reefs 33:1165–1165
Carpenter KE, Abrar M, Aeby G, Aronson RB, Banks S, Bruckner A, Chiriboga A, Cortés J, Delbeek JC, DeVantier L (2008) One-third of reef-building corals face elevated extinction risk from climate change and local impacts. Science 321:560–563
Chen Y-S, Qiu X-B (2013) Ubiquitin at the crossroad of cell death and survival. Chinese Journal of Cancer 32:640
Chomczynski P (1993) A reagent for the single-step simultaneous isolation of RNA, DNA and proteins from cell and tissue samples. Biotechniques 15(532–534):536–537
Cunning R, Bay R, Gillette P, Baker A, Traylor-Knowles N (2018) Comparative analysis of the Pocillopora damicornis genome highlights role of immune system in coral evolution. Scientific Reports 8:16134
Deveraux QL, Takahashi R, Salvesen GS, Reed JC (1997) X-linked IAP is a direct inhibitor of cell-death proteases. Nature 388:300–304
Domart-Coulon I, Tambutté S, Tambutté E, Allemand D (2004) Short term viability of soft tissue detached from the skeleton of reef-building corals. Journal of Experimental Marine Biology and Ecology 309:199–217
Fordyce AJ, Camp EF, Ainsworth TD (2017) Polyp bailout in Pocillopora damicornis following thermal stress. F1000Research 6
Gavish AR, Shapiro OH, Kramarsky-Winter E, Vardi A (2018) Microscale tracking of coral disease reveals timeline of infection and heterogeneity of polyp fate. bioRxiv:302778
Grabherr MG, Haas BJ, Yassour M, Levin JZ, Thompson DA, Amit I, Adiconis X, Fan L, Raychowdhury R, Zeng Q (2011) Trinity: reconstructing a full-length transcriptome without a genome from RNA-Seq data. Nature Biotechnology 29:644
Hobmayer B, Snyder P, Alt D, Happel CM, Holstein TW (2001) Quantitative analysis of epithelial cell aggregation in the simple metazoan Hydra reveals a switch from homotypic to heterotypic cell interactions. Cell and Tissue Research 304:147–157
Hoegh-Guldberg O (1999) Climate change, coral bleaching and the future of the world’s coral reefs. Marine and Freshwater Research 50:839–866
Huang DW, Sherman BT, Lempicki RA (2008) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Research 37:1–13
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protocols 4:44
Hughes DT, Sperandio V (2008) Inter-kingdom signalling: communication between bacteria and their hosts. Nature Reviews Microbiology 6:111–120
Johnston EC, Forsman ZH, Flot J-F, Schmidt-Roach S, Pinzón JH, Knapp IS, Toonen RJ (2017) A genomic glance through the fog of plasticity and diversification in Pocillopora. Scientific Reports 7:5991
Jones R (2005) The ecotoxicological effects of Photosystem II herbicides on corals. Marine Pollution Bulletin 51:495–506
Jones RJ, Hoegh-Guldberg O (1999) Effects of cyanide on coral photosynthesis: implications for identifying the cause of coral bleaching and for assessing the environmental effects of cyanide fishing. Marine Ecology Progress Series 177:83–91
Karin M, Lin A (2002) NF-κB at the crossroads of life and death. Nature Immunology 3:221
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biology 14:R36
Kojima S-i, Itoh Y, Matsumoto S-i, Masuho Y, Seiki M (2000) Membrane-type 6 matrix metalloproteinase (MT6-MMP, MMP-25) is the second glycosyl-phosphatidyl inositol (GPI)-anchored MMP. FEBS Letters 480:142–146
Krejci P, Masri B, Fontaine V, Mekikian PB, Weis M, Prats H, Wilcox WR (2005) Interaction of fibroblast growth factor and C-natriuretic peptide signaling in regulation of chondrocyte proliferation and extracellular matrix homeostasis. Journal of Cell Science 118:5089–5100
Kvitt H, Rosenfeld H, Zandbank K, Tchernov D (2011) Regulation of apoptotic pathways by Stylophora pistillata (Anthozoa, Pocilloporidae) to survive thermal stress and bleaching. PloS One 6:e28665
Kvitt H, Kramarsky-Winter E, Maor-Landaw K, Zandbank K, Kushmaro A, Rosenfeld H, Fine M, Tchernov D (2015) Breakdown of coral colonial form under reduced pH conditions is initiated in polyps and mediated through apoptosis. Proceedings of the National Academy of Sciences 112:2082–2086
Leitão AL, Enguita FJ (2016) Non-coding RNAs and inter-kingdom Communication. Springer
Lesser M, Stochaj W, Tapley D, Shick J (1990) Bleaching in coral reef anthozoans: effects of irradiance, ultraviolet radiation, and temperature on the activities of protective enzymes against active oxygen. Coral Reefs 8:225–232
Li B, Dewey CN (2011) RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics 12:323
Littman R, Willis BL, Bourne DG (2011) Metagenomic analysis of the coral holobiont during a natural bleaching event on the Great Barrier Reef. Environmental Microbiology Reports 3:651–660
Liu J-F, Crépin M, Liu J-M, Barritault D, Ledoux D (2002) FGF-2 and TPA induce matrix metalloproteinase-9 secretion in MCF-7 cells through PKC activation of the Ras/ERK pathway. Biochemical and Biophysical Research Communications 293:1174–1182
Liu C, Cheng SH, Lin S (2020) Illuminating the dark depths inside coral. Cellular Microbiology 22:e13122
Marshall A, Clode P (2004) Effects of calcium-free and low-calcium artificial seawater on polyps of a scleractinian coral Galaxea fascicularis. Coral Reefs 23:277–280
Pacheco AR, Sperandio V (2009) Inter-kingdom signaling: chemical language between bacteria and host. Current Opinion in Microbiology 12:192–198
Pintucci G, Yu PJ, Sharony R, Baumann FG, Saponara F, Frasca A, Galloway AC, Moscatelli D, Mignatti P (2003) Induction of stromelysin-1 (MMP-3) by fibroblast growth factor-2 (FGF-2) in FGF-2 −/− microvascular endothelial cells requires prolonged activation of extracellular signal-regulated kinases-1 and-2 (ERK-1/2). Journal of Cellular Biochemistry 90:1015–1025
Pinzon JH, LaJeunesse TC (2011) Species delimitation of common reef corals in the genus Pocillopora using nucleotide sequence phylogenies, population genetics and symbiosis ecology. Molecular Ecology 20:311–325
Poquita-Du RC, Quek ZBR, Jain SS, Schmidt-Roach S, Tun K, Heery EC, Chou LM, Todd PA, Huang D (2019) Last species standing: loss of Pocilloporidae corals associated with coastal urbanization in a tropical city state. Marine Biodiversity:1-15
Puverel S, Tambutte E, Zoccola D, Domart-Coulon I, Bouchot A, Lotto S, Allemand D, Tambutte S (2005) Antibodies against the organic matrix in scleractinians: a new tool to study coral biomineralization. Coral Reefs 24:149–156
Quistad S, Traylor-Knowles N (2016) Precambrian origins of the TNFR superfamily. Cell Death Discovery 2:1–6
Quistad SD, Stotland A, Barott KL, Smurthwaite CA, Hilton BJ, Grasis JA, Wolkowicz R, Rohwer FL (2014) Evolution of TNF-induced apoptosis reveals 550 My of functional conservation. Proceedings of the National Academy of Sciences:201405912
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Roulston A, Reinhard C, Amiri P, Williams LT (1998) Early activation of c-Jun N-terminal kinase and p38 kinase regulate cell survival in response to tumor necrosis factor α. Journal of Biological Chemistry 273:10232–10239
Sammarco PW (1982) Polyp bail-out: an escape response to environmental stress and a new means of reproduction in corals. Marine Ecology Progress Series Oldendorf 10:57–65
Schmidt-Roach S, Miller KJ, Lundgren P, Andreakis N (2014) With eyes wide open: a revision of species within and closely related to the Pocillopora damicornis species complex (Scleractinia; Pocilloporidae) using morphology and genetics. Zoological Journal of the Linnean Society 170:1–33
Segovia M (2008) Programmed cell death in dinoflagellates Programmed Cell Death in Protozoa. Springer, pp126-142
Serrano E, Coma R, Inostroza K, Serrano O (2018) Polyp bail-out by the coral Astroides calycularis (Scleractinia, Dendrophylliidae). Marine Biodiversity 48:1661–1665
Shapiro OH, Kramarsky-Winter E, Gavish AR, Stocker R, Vardi A (2016) A coral-on-a-chip microfluidic platform enabling live-imaging microscopy of reef-building corals. Nature Communications 7:10860
Sharp KH, Ritchie KB (2012) Multi-partner interactions in corals in the face of climate change. The Biological Bulletin 223:66–77
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM (2015) BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31:3210–3212
Traylor-Knowles N, Granger BR, Lubinski TJ, Parikh JR, Garamszegi S, Xia Y, Marto JA, Kaufman L, Finnerty JR (2011) Production of a reference transcriptome and transcriptomic database (PocilloporaBase) for the cauliflower coral, Pocillopora damicornis. BMC Genomics 12:585
Van Antwerp DJ, Martin SJ, Kafri T, Green DR, Verma IM (1996) Suppression of TNF-α-induced apoptosis by NF-κB. Science 274:787–789
Van Roy F, Berx G (2008) The cell-cell adhesion molecule E-cadherin. Cellular and Molecular Life Sciences 65:3756–3788
Voolstra CR, Li Y, Liew YJ, Baumgarten S, Zoccola D, Flot J-F, Tambutté S, Allemand D, Aranda M (2017) Comparative analysis of the genomes of Stylophora pistillata and Acropora digitifera provides evidence for extensive differences between species of corals. Scientific Reports 7:17583
Webster N, Negri A, Flores F, Humphrey C, Soo R, Botté E, Vogel N, Uthicke S (2013) Near-future ocean acidification causes differences in microbial associations within diverse coral reef taxa. Environmental Microbiology Reports 5:243–251
Wecker P, Lecellier G, Guibert I, Zhou Y, Bonnard I, Berteaux-Lecellier V (2018) Exposure to the environmentally-persistent insecticide chlordecone induces detoxification genes and causes polyp bail-out in the coral P. damicornis. Chemosphere 195:190–200
Weis VM (2008) Cellular mechanisms of Cnidarian bleaching: stress causes the collapse of symbiosis. Journal of Experimental Biology 211:3059–3066
Whitelock JM, Murdoch AD, Iozzo RV, Underwood PA (1996) The degradation of human endothelial cell-derived perlecan and release of bound basic fibroblast growth factor by stromelysin, collagenase, plasmin, and heparanases. Journal of Biological Chemistry 271:10079–10086
Wiens GD, Glenney GW (2011) Origin and evolution of TNF and TNF receptor superfamilies. Developmental & Comparative Immunology 35:1324–1335
Wild C, Rixen T, Sanchez-Noguera C, Stuhldreier I, Jimenez C, Merico A (2014) Massive coral tissue ablations in reefs of Pacific Costa Rica. Galaxea, Journal of Coral Reef Studies 16:13–14
Acknowledgements
This work was supported by JSPS KAKENHI Grant Number JP18J20226. The authors gratefully acknowledge generous support to the Marine Biophysics Unit from Okinawa Institute of Science and Technology Graduate University (OIST). We thank the OIST DNA Sequencing Section (SQC) for constructing RNA-Seq libraries and performing sequencing and the OIST Scientific Computing and Data Analysis Section (SCDA) for providing the high-performance computing service for the transcriptome assembly in this study. We appreciate the efforts of Dr. Steven D. Aird in editing and commenting on the manuscript.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Additional information
Topic Editor Morgan S. Pratchett
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary material 1 (MP4 468 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Chuang, PS., Mitarai, S. Signaling pathways in the coral polyp bail-out response. Coral Reefs 39, 1535–1548 (2020). https://doi.org/10.1007/s00338-020-01983-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00338-020-01983-x