Abstract
Male reproductive anomalies are widely distributed among mammals, and male factors are estimated to contribute to approximately 50% of cases of human infertility. The B10.M/Sgn (B10.M) mouse strain exhibits two adverse reproductive phenotypes: severe teratospermia and male subfertility. Although teratospermia is known to be heritable, the relationship between teratospermia and male subfertility has not been well characterized. The fertility of B10.M male mice is considerably lower (~ 30%) than that of standard laboratory mouse strains (~ 70%). To genetically analyze male subfertility, F2 males were produced by intercrossing the F1 progeny of female B10.M and male C3H/HeN mice. The fertility of each F2 male mouse was assessed based on the outcomes of matings with five females. Statistical analysis of correlations between the two reproductive phenotypes (teratospermia and subfertility) in F2 males (n = 177) revealed that teratospermia is not the cause of male subfertility. Quantitative trait loci (QTL) analysis of the male subfertility phenotype (n = 128) using GigaMUGA markers mapped one significant QTL peak to chromosome 4 at 62.9 centimorgans (cM) with a logarithm of odds score of 11.81 (P < 0.05). We named the QTL locus Mfsf1 (male factor subfertility 1). Further genetic analysis using recombinant males restricted the physical area to 1.53 megabasepairs (Mbp), encompassing 22 protein-coding genes. In addition, we found one significant QTL and one indicative QTL on chromosome 5 and 12, respectively, that interacted with the Mfsf1 locus. Our results demonstrate that genetic dissection of male subfertility in the B10.M strain is a useful model for characterizing the complex genetic mechanisms underlying reproduction and infertility.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Human infertility is a common problem worldwide, and male factors are estimated to contribute to 50% of cases (Agarwal et al. 2015; Melodie and Christine 2018). The etiology of male infertility is highly variable and has been associated with endocrinological, immunological, neurogenic, and environmental factors (Iammarrone et al. 2003). A correlation between male infertility and emergence of cancer has also been reported (Nagirnaja et al. 2018). Although genetic contribution to male factor infertility are evident in humans, limited associations with chromosomal alterations, such as Yq microdeletions and Klinefelter’s syndrome, and mutations in specific genes, such as CFTR, have been reported (Neto et al. 2016). Studies in mice have identified 666 genes that cause male infertility when disrupted (Nagirnaja et al. 2018). As genomic information continues to accumulate, an increasing number of genetic variations linked to male infertility are expected to be identified, and the search for novel genetic variations that underlie male factor infertility is already underway (Miyamoto et al. 2017; Robay et al. 2018; Halder et al. 2017).
Subfertility is defined as reproductive efficiency that lies between that of infertility and normal reproductive performance. This low efficiency is a major concern in agriculture and animal production. For example, in dairy cows, a trend of declining fertility persists for several decades (LeBlanc 2010). Substantial attention has been paid to understand the correlation between milk production and cow fertility (Jamuna and Chakravarty 2016; Berry et al. 2016). With the advent of newly developed technologies in genome science, application of genomic information to improve fertility has become a promising mechanism to improve unfavorable reproductive conditions (Taylor et al. 2018).
Use of model mouse strains to characterize fertility is advantageous because the effects of genetic background can be assessed in a reproducible manner. For example, the well-characterized B10.M/Sgn mouse strain is characterized by male subfertility and severe teratospermia, with ~ 60% of sperm exhibiting abnormal morphology (Gotoh 2010). Previously, we reported that the teratospermia phenotype is heritable (Gotoh et al. 2012) and mapped two of the loci responsible: Shm1 on chromosome 1 and Shm2 on chromosome 4. Interactions between these two loci were evident. Homozygosity of the Shm1B10.M allele was required to express teratospermia, and the enhancing effects of the Shm2B10.M/B10.M genotype were observed only when the genotype of the animal was homozygous for the Shm1B10.M allele. However, whether a link exists between the teratospermia phenotype and male subfertility in this mouse strain has not previously been investigated. Therefore, in the present study, we performed genetic analyses on the male subfertility phenotype and explored linkage with the teratospermia phenotype.
Materials and methods
Animals
All experiments were approved by the Institutional Animal Care and Use Committee of the Institute of Agrobiological Sciences. Animals were housed and cared for according to guidelines established by the Committee. C3H/HeNCrlCrlj (C3H) mice were purchased from Charles River Japan (Yokohama, Japan). C57BL/10J (B10) mice were purchased from S. L. C. (Hamamatsu, Japan). B10.M/Sgn (B10.M) mice are maintained at our facility. Animals were maintained on a cycle of 12 h of light and 12 h of darkness under specific-pathogen-free conditions. The commercial mouse diet CRF-1 (Charles River Japan, Yokohama, Japan) and water were provided. F2 animals were produced by intercrossing F1 animals obtained from crossing B10.M females with C3H males. Recombinant F2 males in which recombination occurred between the D4Mit251, D4Mit54, and D4Mit170 microsatellite markers were selected for fine mapping of the locus responsible for the male subfertility phenotype detected by QTL analysis.
Male fertility analysis
Mature, virgin, C3H females ages 8–12 weeks were used for the assay. Five female mice were mated with each male mouse. Male fertility is assessed according to a standard methodology for reproductive toxicological assay (Teramoto et al. 1980; Mitchard et al. 2012). Two weeks after the vaginal plug was observed, the female mice were dissected. The numbers of corpus luteum (yellow body) on ovaries and the numbers of embryos in uterus were counted under a stereo microscope (SMZ25; Nikon, Tokyo). Because yellow body is the remains of ovarian follicle that has released a mature ovum during a previous ovulation, the ratio of the total number of embryos to the number of yellow bodies was used to estimate the fertility percentage for each mating. The fertility of each male mouse was expressed as the mean fertility percentage for the five matings.
Sperm morphology test
Sperm samples were collected from 3- to 5-month-old male mice after fertility testing was completed. Sperm morphology was analyzed as described earlier (Gotoh 2010). Two independent samples, each containing a minimum of 200 sperm cells, were analyzed under 400 × magnification using a differential interference contrast microscope (DMRXA2; Leica Microsystems; Cambridge, UK).
Quantitative trait loci (QTL) analysis
Genomic DNA samples were prepared from mouse tail snips (~ 5 mm) as previously described (Hirawatari et al. 2015b). Samples from 128 F2 animals were genotyped using GigaMUGA markers (Morgan et al. 2015) at the Genetics Laboratory, University of North Carolina, Chapel Hill. QTL analysis was performed using R/qtl software (Broman et al. 2003; Broman and Sen 2009). The scanone function was used for single QTL analysis, and the scaonetwo function was used to detect epistatic interactions between two loci. The threshold for each assay was determined by permutation tests (n = 1000). P values less than 0.05 and 0.37 were considered statistically significant and suggestive, respectively, for QTL analyses.
Database search
Information of position, MGI ID, feature type, and symbol of genes is referred to the Mouse Genome Informatics (MGI) website (http://www.informatics.jax.org/marker). Information of known reproductive phenotype of genes is referred to the International Mouse Phenotyping Consortium (IMPC) website (http://www.mouse.phenotype.org). Information of expression of genes in testis is referred to the Expression Atlas database of the European Bioinformatics Institute website (http://www.ebi.ac.uk/gxa.home). Information of single nucleotide polymorphism (SNP) of genes among C3H/He, C3H/HeJ, C57BL/10J, C57BL/6, and C57BL6J inbred strains is referred to the SNP database of the MGI website (http://www.informatics.jax.org/snp). All information was updated on July 31, 2018.
Statistical analyses
Correlations between male subfertility and teratospermia phenotypes were analyzed using Spearman’s rank correlation test. Comparisons between groups were analyzed by one-way analysis of the variance (ANOVA) using SPSS 16.0 for Windows (Analytical Software; Chicago, IL, USA).
Results
Male subfertility and teratospermia
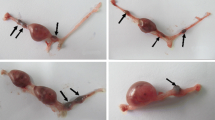
Figure 1a shows a representative example of the ovaries and a uterus from a C3H female mated with a sub-fertile F2 male. In this case, nine yellow bodies and two embryos were present, resulting in a fertility estimate of 22%. Figure 1b presents an example of a sperm spread from an F2 male exhibiting severe teratospermia (64% abnormal spermatozoa) with a number of morphological abnormalities.
Abnormal male reproductive phenotypes present in the B10.M strain. a A representative example of the fertility assay used in this study showing subfertility of an F2 male. Upper photos show a pair of ovaries. The right (R) ovary contains eight yellow bodies, and the left (L) ovary contains one yellow body. Horizontal bar, 100 μm. Lower photo shows a uterus containing fetuses on 13th day of pregnancy. Each side of the uterus contains one fetus, respectively. The fertility was calculated to be 22% in this case. Horizontal bar, 10 mm. b An example of teratospermia of an F2 male. Abnormal sperm head morphology is shown. n represents a normal spermatozoon; a shows ectopic attachment of the flagellum; a shows a spermatozoon with amorphous head, and l shows the lack of the usual hook
Male subfertility is not correlated with teratospermia in mice
The correlation between male fertility and sperm morphological abnormalities in F2 males is shown in Fig. 2. The horizontal dotted line at 58.7% represents the statistical mean fertility minus the standard deviation (SD) for B10 males (n = 10). The vertical-dotted line at 9.9% represents the statistical mean percentage of sperm shape abnormalities plus the SD for B10 males (n = 10). These lines define approximate boundaries between high and low values for each phenotype. A considerable number of individuals are found in both the upper-right square and the lower-left quadrants. Males plotted in the upper-right quadrant exhibit normal fertility with teratospermia. Males plotted in the lower-left quadrant exhibit subfertility with normal sperm morphology. Statistical analysis of F2 males (n = 177) confirmed the lack of a correlation between the two phenotypes (P > 0.05).
Dot matrix evaluating the relationship between teratospermia and male subfertility phenotypes. Each dot represents the sperm shape abnormality and fertility scores for one F2 male. The horizontal-dotted line indicates a rough threshold to differentiate between normal and low fertility (58.7%; mean − SD). The vertical-dotted line indicates a rough threshold to differentiate between normal and high frequencies of abnormal sperm morphology (9.9%; mean + SD). A statistically significant correlation was not identified by Spearman’s correlation test (P > 0.05)
QTL mapping of a male factor subfertility locus
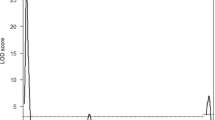
The scanone function of the R/qtl package revealed one significant QTL peak on chromosome 4 at 62.94 cM with an LOD score of 11.81 (Fig. 3a). We tentatively named this locus male factor subfertility 1 (Mfsf1). These results confirmed that male subfertility is heritable in the B10.M strain. The marker nearest the QTL peak is UNC8352858. The effects of the UNC8352858 genotype on male fertility are shown in Fig. 3b. Chromosome interval mapping using the R/qtl package estimated that the Mfsf1 locus mapped between the UNCHS013009 (59.84 cM) and UNCHS013136 (63.47 cM) markers (P < 0.05).
QTL analysis of the male subfertility phenotype in the B10.M strain. a One significant peak (LOD score, 11.81) is located on chromosome 4 at 62.94 cM. The horizontal solid line (LOD score, 3.97) and dotted line (LOD score, 2.83) indicate thresholds for statistical significance (P > 0.05) and suggestiveness (P > 0.37), respectively. The LOD score for the QTL peak identified on chromosome 12 is 2.72. The thresholds were determined by permutation testing (n = 1000). b Effect of UNC8252838 genotype on male fertility. The marker nearest the QTL peak on chromosome 4 is UNC8352838. Each circle indicates the fertility of a male. The mean fertility ± standard error for each genotype is shown to the right of the circles. AA indicates homozygosity for the B10.M allele, AB indicates heterozygosity, and BB indicates homozygosity for the C3H allele
Genetic mapping of the Mfsf1 locus
Genetic mapping of the Mfsf1 locus was performed using conventional microsatellite markers and recombinant males to narrow the mapping region obtained by QTL analysis. QTL results indicated that homozygosity of the Mfsf1B10.M allele was required to express the subfertility phenotype, although the fertility values of the Mfsf1B10.M/B10.M homozygotes varied widely (Fig. 3b). Three informative recombinant males were obtained. The Mfsf1 locus was mapped between the D4Mit251 and D4Mit170 markers by this analysis (Fig. 4a). Together with QTL mapping results, these results indicate that the Mfsf1 locus is restricted to a 1.53 Mbp region containing 22 protein-coding genes (Fig. 4b). Known information of candidate genes within the mapped interval is listed in Supplemental Table S1.
Genetic mapping of the Mfsf1 locus within the QTL interval on chromosome 4 using recombinant males. a Genotypes M and H indicate homozygosity and heterozygosity of the B10.M allele, respectively. The pink and the light green regions originated from B10.M chromosomes indicate the presence and absence of the Mfsf1 locus, respectively. The observed fertility and the identification number of the animal are listed to the right. b Comparison of mapping of the Mfsf1 locus by QTL and genetic analyses. Marker positions refer to the UniSTS annotation for GRCm38. Known protein coding genes within the mapped area are listed at the bottom
Epistatic interactions with the Mfsf1 locus
The wide distribution of fertility in Mfsf1B10.M/B10.M homozygotes could not be explained by Mfsf1 genotype alone. No significant QTL peaks showing additive effects were found. Therefore, we next searched for other factors that could interact with the Mfsf1 locus using the scantwo function of the R/qtl package. One significant interacting QTL peak was found on chromosome 5 at 40.13 cM; this locus had a suppressive effect on fertility (Fig. 5a). In addition, one QTL peak with an enhancing effect on fertility was found on chromosome 12 at 20.89 cM. The LOD score for this single QTL peak (2.72) was slightly below the suggestive level (Fig. 5b). No significant interaction of this factor with the Mfsf1 locus was found.
Epistatic effects of factors genetically interacting with the Mfsf1 locus on fertility. The UNC8352858 marker represents the nearest marker to the Mfsf1 locus on chromosome 4. a Interaction plot for UNC8352858 and UNCHS015145 on chromosome 5. Homozygosity of the UNCHS025145B10.M allele suppresses fertility. b Interaction plot for UNC8352858 and UNC21089226 on chromosome 12. Homozygosity of the UNC21089226B10.M allele enhances fertility. AA indicates homozygosity for the B10.M allele, AB indicates heterozygosity, and BB indicates homozygosity for the C3H allele
Discussion
This study revealed that male subfertility in the B10.M mouse strain is heritable. The Mfsf1 locus responsible for subfertility is mapped to chromosome 4. It is different from the major locus (Shm1) responsible for teratospermia on chromosome 1 (Hirawatari et al. 2015b). We also found that the subfertility phenotype was not associated with the previously reported sperm shape abnormality phenotype in B10.M strain. Similarly, analysis of the TEN1 mouse strain also reported that teratospermia was not associated with male subfertility (Hirawatari et al. 2015a). Male subfertility is often attributed to teratospermia. The results of this study clearly show that sperm shape abnormality phenotype and male subfertility phenotype are not always linked.
Although male subfertility and teratospermia are not associated in B10.M mice, this does not necessarily imply that there is no association between the genes responsible for these two phenotypes. With respect to the teratospermia phenotype in B10.M mice, the two loci responsible, Shm1 on chromosome 1 and Shm2 on chromosome 4, have been shown to interact (Gotoh et al. 2012). Homozygosity of the Shm1B10.M allele is required for expression of the sperm shape phenotype, and the Shm2B10.M allele enhances the frequency of morphologically abnormal sperm in a recessive manner. The Shm2B10.M allele alone does not cause teratospermia. The Shm2 locus has been mapped to chromosome 4 between the D4Mit148 (69.48 cM, 137148689 bp) and D4Mit170 (70.47 cM, 138615338 bp) markers. Thus, the two mapped regions, the Shm2 locus in the teratospermia study and the Mfsf1 locus in this study, are nearly identical, and the genes encoded by the Shm2 locus and the Mfsf1 locus could possibly be identical. Only 22 protein-coding genes are located within the restricted mapped area (Supplemental Table S1).
Because the B10.M strain is an h2-congenic strain possessing the h2-complex on chromosome 17 in a genetic background derived 75% from C57BL/6 (B6) and 25% from B10 strains (Gotoh 2010), genes on chromosome 4 have originated from either B6 or B10. By searching SNP database, several non-synonymous SNP variants between C57BL/6J and C3H/HeJ strains are found within the coding sequences of the candidate genes (Supplemental Table S2). SNP information of B10 strain for these genes is not available. Because neither B10 males nor C3H males show subfertile phenotype (Hirawatari et al. 2015b), the listed SNP variation is unlikely to cause subfertility of B10.M. A novel mutation appears to have emerged during or after establishment of the B10.M inbred strain. With recent advances in technology, whole genome DNA sequencing is now available for mouse, and genome editing technology has become convenient. These techniques will be required to find genetic variations within the mapped region and then show that the subfertility phenotype can be induced by the identified mutation.
Epistasis, or interaction between genes, was observed in this study. Homozygosity of the Mfsf1B10.M allele appears to be required to express male subfertility in the B10.M strain, as the other two genotypes, heterozygotes and wild-type homozygotes, exhibit normal fertility. However, less than half of the Mfsf1B10.M/B10.M homozygous males displayed the subfertility phenotype (Fig. 3b). The QTL analysis in this study indicated two bidirectional factors present on chromosomes 5 and 12 that interacted with the Mfsf1 locus. The locus on chromosome 12 was the only factor found to enhance fertility. Likely due to the limited sample size in this study, the interacting effect of the QTL on chromosome 12 was not statistically significant.
Results from this study demonstrate that genetic approaches can be a powerful tool to analyze mechanisms underlying the complexity of the reproductive system. These approaches are useful as a large number of the 3000 genes expressed in male germ cells may contribute to male infertility (Schultz et al. 2003). In fact, gene knockout studies in mice have already identified 666 genes associated with male infertility (Nagirnaja et al. 2018). In addition, genetic interactions associated with reproductive phenotypes are common. Over the course of evolutionary history, each mammalian species or subspecies has acquired complex mechanisms to regulate normal reproduction. Haldane’s rule in mammals (Coyne 1985) and the hybrid sterility observed between two Mus musculus (house mouse) subspecies (Storchova et al. 2004) are two examples of these types of regulatory mechanisms. Another is typified by the frequent appearance of infertile animals among the hybrid descendants of two reproductively normal, inbred mouse strains (Nishimura et al. 1995; Shorter et al. 2017). However, reproductive regulatory mechanisms are not only defined by the extreme condition of infertility, as a wide variety of male reproductive phenotypes has been observed among F2 hybrid males produced by reproductively normal B6 and C3H strains in the absence of a novel mutation (Gotoh and Aoyama 2012). In the genetic study of teratospermia in the TEN1 mouse strain, at least three interacting loci, Shm3 on chromosome 1, Shm4 on X chromosome, and Shm5 on chromosome 6, have been identified (Hirawatari et al. 2015b). In cases in which a single genetic variation causes infertility, genomic methods can be powerful tools for finding the causative gene. However, male reproductive system is supposed to be composed of multiple interactions of genes.
References
Agarwal A, Mulgund A, Hamada A, Chyatte MR (2015) A unique view on male infertility around the globe. Reprod Biol Endocrinol 13:37
Berry DP, Friggens NC, Lucy M, Roche JR (2016) Milk production and fertility in cattle. Annu Rev Anim Biosci 4:269–290
Broman KW, Sen S (2009) A guide to QTL mapping with R/qtl. Springer, New York
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Coyne JA (1985) The genetic basis of Haldane’s rule. Nature 314:736–738
Gotoh H (2010) Inherited sperm head abnormalities in the B10.M mouse strain. Reprod Fertil Dev 22:1066–1073
Gotoh H, Aoyama H (2012) Spermatogenic defects in F2 mice between normal mouse strains C3H and C57BL/6 without mutation. Congenit Anom (Kyoto) 52:186–190
Gotoh H, Hirawatari K, Hanzawa N, Miura I, Wakana S (2012) QTL on mouse chromosomes 1 and 4 causing sperm-head morphological abnormality and male subfertility. Mamm Genome 23:399–403
Halder A, Kumar P, Jain M, Kalsi AK (2017) Genomics: tool to predict and prevent male infertility. Front Biosci (Schol ed) 9:448–508
Hirawatari K, Hanzawa N, Kuwahara M, Aoyama H, Miura I, Wakana S, Gotoh H (2015a) Polygenic expression of teratozoospermia and normal fertility in B10.MOL-TEN1 mouse strain. Congenit Anom (Kyoto) 55:92–98
Hirawatari K, Hanzawa N, Miura I, Wakana S, Gotoh H (2015b) A cascade of epistatic interactions regulating teratozoospermia in mice. Mamm Genome 26:248–256
Iammarrone E, Balet R, Lower AM, Gillott C, Grudzinskas JG (2003) Male infertility. Best practice & research. Clin Obstet Gynaecol 17:211–229
Jamuna V, Chakravarty AK (2016) Evaluation of fertility in relation to milk production and productivity of Murrah buffaloes. Anim Reprod Sci 171:72–80
LeBlanc S (2010) Assessing the association of the level of milk production with reproductive performance in dairy cattle. J Reprod Dev 56(Suppl):S1–S7
Melodie VB, Christine W (2018) Fertility and infertility: definition and epidemiology. Clin Biochem. https://doi.org/10.1016/j.clinbiochem.2018.03.012
Mitchard T, Larvis P, Stewart J (2012) Assessment of male rodent fertility in general toxicology 6-month studies. Birth Defects Res B Dev Reprod Tocicol 95:410–420
Miyamoto T, Minase G, Shin T, Ueda H, Okada H, Sengoku K (2017) Human male infertility and its genetic causes. Reprod Med Biol 16:81–88
Morgan AP, Fu CP, Kao CY, Welsh CE, Didion JP, Yadgary L, Hyacinth L, Ferris MT, Bell TA, Miller DR, Giusti-Rodriguez P, Nonneman RJ, Cook KD, Whitmire JK, Gralinski LE, Keller M, Attie AD, Churchill GA, Petkov P, Sullivan PF, Brennan JR, McMillan L, Pardo-Manuel de Villena F (2015) The mouse universal genotyping array: from substrains to subspecies. G3 (Bethesda) 6:263–279
Nagirnaja L, Aston KI, Conrad DF (2018) Genetic intersection of male infertility and cancer. Fertil Steril 109:20–26
Neto FT, Bach PV, Najari BB, Li PS, Goldstein M (2016) Genetics of male infertility. Curr Urol Rep 17:70
Nishimura M, Hirayama N, Serikawa T, Kanehira K, Matsushima Y, Katoh H, Wakana S, Kojima A, Hiai H (1995) The SMXA: a new set of recombinant inbred strain of mice consisting of 26 substrains and their genetic profile. Mamm Genome 6:850–857
Robay A, Abbasi S, Akil A, El-Bardisi H, Arafa M, Crystal RG, Fakhro KA (2018) A systematic review on the genetics of male infertility in the era of next-generation sequencing. Arab J Urol 16:53–64
Schultz N, Hamra FK, Garbers DL (2003) A multitude of genes expressed solely in meiotic or postmeiotic spermatogenic cells offers a myriad of contraceptive targets. Proc Natl Acad Sci USA 100:12201–12206
Shorter JR, Odet F, Aylor DL, Pan W, Kao CY, Fu CP, Morgan AP, Greenstein S, Bell TA, Stevans AM, Feathers RW, Patel S, Cates SE, Shaw GD, Miller DR, Chesler EJ, McMillian L, O’Brien DA, Villena FP (2017) Male infertility is responsible for nearly half of the extinction observed in the mouse collaborative cross. Genetics 206:557–572
Storchova R, Gregorova S, Buckiova D, Kyselova V, Divina P, Forejt J (2004) Genetic analysis of X-linked hybrid sterility in the house mouse. Mamm Genome 15:515–524
Taylor JF, Schnabel RD, Sutovsky P (2018) Identification of genomic variants causing sperm abnormalities and reduced male fertility. Anim Reprod Sci 194:57–62
Teramoto S, Saito R, Aoyama H, Shirasu Y (1980) Dominant lethal mutation induced in male rates by 1,2-dibromo-3-chloropropane (DBCP). Mutat Res 77:71–78
Acknowledgements
The authors would like to thank Mr. Heiichi Uchiyama for his assistance with the experiments. We also thank BioMed Proofreading® LLC for proofreading of the manuscript. This work was supported by the Ministry of Agriculture, Forestry, and Fisheries, Japan.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Gotoh, H., Miura, I. & Wakana, S. Genetic mapping of a male factor subfertility locus on mouse chromosome 4. Mamm Genome 29, 663–669 (2018). https://doi.org/10.1007/s00335-018-9773-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-018-9773-4