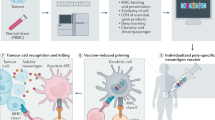
Abstract
This review focuses on the types of cancer antigens that can be recognised by the immune system and form due to alterations in the cancer genome, including cancer testis, overexpressed and neoantigens. Specifically, neoantigens can form when cancer cell-specific mutations occur that result in alterations of the protein from ‘self’. This type of antigen can result in an immune response sufficient to clear tumour cells when activated. Furthermore, studies have reported that the likelihood of successful immunotherapeutic targeting of cancer by many different methods was reliant on immune response to neoantigens. The recent resurgence of interest in the immune response to tumour cells, in conjunction with technological advances, has resulted in a large increase in the predicted, identified and functionally confirmed neoantigens. This growth in identified neoantigen sequences has increased the contents of training sets for algorithms, which in turn improves the prediction of which genetic mutations may form neoantigens. Additionally, algorithms predicting how proteins will be processed into peptide epitopes by the proteasome and which peptides bind to the transporter complex are also improving with this research. Now that large screens of all the tumour-specific protein altering mutations are possible, the emerging data from assessment of the immunogenicity of neoantigens suggest that only a minority of variants will form targetable epitopes. The potential for immunotherapeutic targeting of neoantigens will therefore be greater in cancers with a higher frequency of protein altering somatic variants. There is considerable potential in the use of neoantigens to treat patients, either alone or in combination with other immunotherapies and with continued advancements, these potentials will be realised.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
While the immune system has been known to play a role in the control of tumourigenic cells since the start of the twentieth century (historical review: Strebhardt and Ullrich 2008), research has yet to identify consistent methods to manipulate it to clear tumour cells. Significant technological advances have allowed researchers to molecularly characterise tumours and responding immune cells, which has resulted in breakthroughs that have translated to pharmacologically actionable markers and targets (as reviewed in Pritchard 2018). In order to reduce potential side effects to the patient, markers that are unique to cancer cells are particularly desirable. This review will focus on the immunogenic antigens that are expressed by cancer cells, the methods by which a particular type of antigen can be identified from genomic data and what is understood about the immunogenic potential of these antigens. Immune cells that can recognise cancer cells displaying antigen markers that are specific to tumour cells include CD4+ and CD8+ T-cells and B-cell subsets. This review only examines the immune targets displayed by cancer cells that are recognised by T-cells; a recommended review of the role of B-cells is Yuen et al. (2016).
Immune recognition of antigens
In order to be displayed on cells, these antigens have to go through a process of protein cleavage and binding to MHC molecules; then in order to be recognised by the T-cell, a T-cell receptor (TCR) capable of binding the displayed peptide/MHC complex (pMHC) must be present. These processes are outlined in the following section.
The major histocompatibility complex
The MHC (major histocompatibility complex) is expressed nearly ubiquitously on the majority of cells in vertebrates. The MHC displays protein fragments sampled from both within and outside the cell to alert the immune system to infection by pathogens. As the MHC display peptides from all protein sources, the T-cells recognising the MHC/peptide complexes have to be able to distinguish ‘self’ from ‘non-self’ to avoid autoimmunity. In humans, the HLA proteins are encoded by genes that form a cluster on chromosome 6. They are broadly split into two types: MHC-class I and MHC-class II molecules. Humans have three classical MHC-class I genes, called HLA-A, HLA-B and HLA-C, and three classical MHC-class II molecules: HLA-DR, HLA-DQ and HLA-DP; non-classical MHC molecules also exist. Different subtypes of T-cells recognise MHC/peptide complexes, with CD8+ T-cells recognising internally derived peptides bound to MHC-class I and CD4+ T-cells recognising peptides derived from external proteins bound to MHC-class II. A recommended comprehensive review on the structure of the MHC molecules is Blum et al. (2013).
Protein processing by the proteasome
Proteins are processed into peptide fragments by the proteasome (Uebel and Tampe 1999). There are different proteasomes that can generate peptides for MHC-class I presentation, dependent on the cell source (Basler et al. 2013; Kloetzel 2004). The manner by which proteins are cleaved to form short peptides capable of binding to the MHC molecules has been examined. This has largely been based on in-depth assessment of the processing of specific proteins [e.g. enolase by the immunoproteasome (Toes et al. 2001) and β-casein by the 26S proteasome (Emmerich et al. 2000)] and by examining the known peptides produced and bound to MHC molecules, in the context of the whole protein. Despite the relatively few studies examining how whole proteins are processed through the different proteasomes, these data have been used as training sets for development of in silico prediction tools such as NetChop (Kesmir et al. 2002), BP-NN (Wang et al. 2013) and mhc-pathway (Tenzer et al. 2005).
Peptide binding to MHC molecules
After peptides have been processed in the cytosol by the proteasome, they are selected for movement to the endoplasmic reticulum (ER) by their ability to bind the TAP (transporter associated with antigen processing) complex (Lehnert and Tampe 2017). The ability of the peptide to bind to TAP is also an aspect of MHC processing that has been investigated for predictability testing (e.g. Bhasin et al. 2007; Peters et al. 2003; Tenzer et al. 2005; Zhang et al. 2006). Once within the ER, peptides are loaded on to the HLA proteins based on their ability to fit in the binding groove. The HLA/peptide complex is then shuttled to the cell surface in complex with chaperone proteins for display to the immune system.
The nature of the binding of peptides to the MHC molecules and factors that can influence these interactions are discussed in the next sections; for more comprehensive reviews on the structure of the MHC molecules and recognition of the MHC-peptide complex by T-cells, please see Blum et al. (2013) and Wucherpfennig et al. (2010), respectively.
Peptide binding to MHC-class I
X-ray crystallographic structure of the MHC-class I molecule showed that the binding groove is composed of two α-helical regions forming the sides and eight antiparallel β-strands that form its floor (Bjorkman et al. 1987a, b). MHC-class I molecules bind short peptide epitopes, with the N- and C-terminal ends being anchored into pockets at each end of the peptide binding groove (Natarajan et al. 1999). The different MHC-class I HLA subtypes tend to bind specific amino acids in these anchor points. The majority of MHC-class I binding peptides are 9 amino acids long (Fig. 1); however, peptides from 8 to 15 amino acids have been discovered. This is due to ‘bulging’ of the central section of the peptides, which allows the peptide to still fit within the binding groove (Fremont et al. 1992; Guo et al. 1992; Tynan et al. 2005). Furthermore, it has been shown that some pMHC-class I complexes are more immunogenic that others. Prediction models show that the amino acid position of a presented peptide is an important factor in this immunogenicity, where amino acids with large aromatic side chains may be better recognised by T-cells (Calis et al. 2013).
Peptide binding to MHC-class II
The MHC-class II epitope binding site consists of a groove and several pockets; X-ray crystallographic studies have shown these structures are provided by a β-sheet and two α-helices (Stern et al. 1994; Zhu et al. 2003). Unlike MHC-class I, the peptides that bind to MHC-class II proteins are not limited by the size of the binding groove, due to the MHC-class II peptide binding groove being open at both ends, allowing the binding of peptides up to 30 amino acids (Nelson and Fremont 1999). This results in different numbered positions within the peptide being able to bind to the anchor residues of MHC-class II molecules and makes residue motifs, rather than anchor positions important in defining and predicting peptides with optimal binding.
MHC-class I/TCR interaction
The T-cell receptor is composed of two membrane anchored polypeptides, α and β that each contain one constant(C) and one variable domain. It is these hypervariable loops that recognise the pMHC complex displayed by cells. The pMHC complexes must form an immunological synapse with a TCR on T-cell in order to induce an immune response, which is dependent on additional co-stimulation and secretion of immunostimulatory cytokines by the activated T-cells to be sustained (Grakoui et al. 1999; Monks et al. 1998; Smith-Garvin et al. 2009; reviewed in Smith-Garvin et al. 2009). Cross-reactivity between TCR and pMHC recognition is likely given the number of possible combinations, which is an important consideration when manipulating TCR for immunotherapeutic purposes (Tan et al. 2015). Large-scale detection of antigen-specific T-cells is now possible using peptide MHC multimer technology (Bentzen et al. 2016; Luimstra et al. 2018), and the production of chimeric antigen receptors (Sharma and Kranz 2016), adoptive transfer of neoepitope reactive T-cells (Prickett et al. 2016) and TCR gene therapy (Blankenstein et al. 2015; Kato et al. 2018; Linnemann et al. 2014) have been shown to be promising treatment modalities utilising in silico prediction models.
Types of cancer antigens
There are 3 broad classifications of tumour antigens that can be recognised as immune targets by T-cells: (a) cancer testis antigens (CT), (b) tumour-associated antigens (TA), and (c) tumour-associated antigens (TAA), including viral antigens (White et al. 2014) [e.g. human papilloma virus on cervical or oropharyngeal cancers (Gillison et al. 2000; Walboomers et al. 1999)] and neoantigens. This review will focus on those arising from genomic changes within cells as part of the tumourigenic process.
CT antigens
CT antigens are a family of tumour-associated epitopes expressed on human tumours, but not on other tissues except for testis and placenta. Epigenetic alteration(s) appear to be the main mechanism regulating CT expression both in normal and neoplastic cells (Karpf and Jones 2002; Zendman et al. 2003). These epigenetic modifications allow for tissue-specific expression of transcripts in differentiated tissues and during development (Baylin and Jones 2011). The first description of epigenetic control of a CT antigen was of MAGE-1 promoter hypomethylation (De Smet et al. 1996, 1999), where demethylation removes the expression silencing mark. Since then, DNA methylation and histone post-translational modifications have been shown to be the most commonly employed mechanism controlling re-expression of the genes encoding CT antigens in tumour cells (Fratta et al. 2011; Siebenkas et al. 2017), reviewed in (Akers et al. 2010). Examples include NY-ESO-1 (Gnjatic et al. 2006) and members of the MAGE family (Chomez et al. 2001); a comprehensive list and evaluation of CT antigens are available at http://www.cta.lncc.br/ (Ludwig Institute for Cancer Research).
CT antigens represent promising therapeutic targets due to the following factors: (a) outside of tumour cells, expression is limited to germ cells (Chen et al. 2009; dos Santos et al. 2000; Greve et al. 2014; Hofmann et al. 2008; Sahin et al. 1998; Zendman et al. 2001); (b) epitopes from CT antigens are not displayed in the testis, as those cells do not express MHC-class I (Fiszer and Kurpisz 1998); (c) as the immune system has not interacted with the CT proteins, it is capable of being recognised as ‘non-self’ (Kalejs and Erenpreisa 2005). Further, immune responses to CT antigens are frequently observed in cancer patients (Akcakanat et al. 2004; Ayyoub et al. 2003; Milne et al. 2008; Qian et al. 2004; Tsuji et al. 2009; Wang et al. 2004) and there is an association between CT antigen expression and activity of tumour immune infiltrates (Rooney et al. 2015).
TA antigens
TA antigens are overexpressed by cancer cells and comprise a large group that broadly can encompass any protein found at increase levels compared with normal tissue. They can be classified as (a) differentiation antigens, which are normal proteins overexpressed as a consequence of the tumourigenic proliferation of cells of a specific function [e.g. pigment production genes, such as tyrosinase in melanomas (Brichard et al. 1993) and the B-cell lineage-specific CD19 (Wang et al. 2012)]; or (b) overexpressed antigens, which are proteins that are minimally expressed by healthy, normal tissues, but are constitutively overexpressed by tumours as part of their malignant phenotype [e.g. PRAME (Kessler et al. 2001), p53 (Barfoed et al. 2000) and ERBB2 in breast cancer (Ellsworth et al. 2008)]. A comprehensive list of these antigens is available in an online database: https://caped.icp.ucl.ac.be/Peptide/list.
As TA antigens are derived from proteins that are overexpressed in a relatively high proportion of a given tumour type, as well as across different cancers, they represent attractive targets for the development of immunotherapy. While a number of antigenic peptides have been reported where immunoreactivity is observed (Vigneron et al. 2013), their use as an immunotherapy target is not devoid of risk. As they are expressed in normal tissue, TA antigens are more likely to have induced immunological tolerance and are less likely to stimulate effective anti-tumour immune responses (Cloosen et al. 2007; Yu et al. 2004).
Types of neoantigen
Neoantigens can arise from any genomic mutation altering protein sequence, including non-synonymous mutations (e.g. Lennerz et al. 2005; Pritchard et al. 2015a), retained introns (e.g. Lupetti et al. 1998), post-translational modification that alters amino acid (e.g. Skipper et al. 1996), gene fusions (e.g. Chang et al. 2017) and frameshift in/del variants (e.g. Inderberg et al. 2017; Linnebacher et al. 2001). Next-generation sequencing (NGS) can be used to identify each of these types of variants, except for post-translational modification, which relies on techniques such as mass spectrometry. As the genomic variations are specific to cancer cells and are not present in the germline, they are not subject to central and peripheral tolerance. First identified in murine models (De Plaen et al. 1988; Monach et al. 1995), neoantigens have subsequently been shown to illicit an immune response capable of clearing tumour (Lennerz et al. 2005; Segal et al. 2008).
Immunotherapeutic potential of cancer antigens
While the above-mentioned tumour antigens have been shown to elicit a robust immune response using autologous and donor in vitro testing (e.g. Jager et al. 1998; Knuth et al. 1984; Lennerz et al. 2005; Murray et al. 1992; Pritchard et al. 2015a; van der Bruggen et al. 1991; Vella et al. 2009), the translation of this to clinical application has largely resulted in low overall response rates (Ilyas and Yang 2015; Neller et al. 2008), with some notable exceptions (e.g. Bollard et al. 2014; Roskrow et al. 1998; Tran et al. 2014, 2016; Zacharakis et al. 2018). As neoantigens are more likely to be different to ‘self’ than CT or TA antigens, the affinity of the TCR recognising the HLA-bound peptide and subsequent strength of the immune response tends to be stronger (Aleksic et al. 2012; Tan et al. 2015). Furthermore, as TA antigens are produced by normal cells and CT antigens may have significant homology to proteins produced on normal cells, their use poses a risk of autoimmunity [e.g. destruction of normal melanocytes in the skin, eye and ear (Johnson et al. 2009)], which can sometimes have devastating unintended consequences to the patient (e.g. Cameron et al. 2013; Morgan et al. 2010, 2013). The more successful clinical approaches have therefore tended to be using viral (e.g. Bollard et al. 2014; Heslop et al. 2010; Louis et al. 2010; Schuessler et al. 2014; Smith et al. 2017) and neoantigen targeting (Sahin et al. 2017; Tran et al. 2014, 2016).
This review will now focus on the neoantigens; for more general overviews of the processes involved in antigen processing and targeting these antigens immunotherapeutically, the following reviews are recommended: Coulie et al. (2014), Lu and Robbins (2016), Pritchard (2018), Tashiro and Brenner (2017).
Identification of neoantigens
Some tumours are more highly mutated than others (Lawrence et al. 2013), resulting in a difference in likelihood of immunogenic neoantigen production between cancer types (Schumacher and Schreiber 2015). As melanoma is among the most genomically mutated tumours, with a high number of non-synonymous single-nucleotide variants (Hayward et al. 2017; Lawrence et al. 2013), it is one of the most frequently studied (e.g. Lauss et al. 2017; Lennerz et al. 2005; Ott et al. 2017; Pasetto et al. 2016; Pritchard et al. 2015a; Snyder et al. 2014; Stronen et al. 2016; van Rooij et al. 2013; Verdegaal et al. 2016). Prior to the advent of massively parallel NGS, the method to identify neoantigens was by labour-intensive individual cDNA library screening (e.g. as performed in Lennerz et al. 2005) and as a result, the number of identified and studied neoantigens was fairly low. Once whole exome/genome sequencing became a routine technique (reviewed in Goodwin et al. 2016), the ability to identify tumour-specific genetic mutations altering the protein coding regions became rapid and high throughput, facilitating neoantigen prediction. Mass spectrometry can also be used to identify peptides bound to the MHC molecules on the surface of cells (Bassani-Sternberg et al. 2016; Freudenmann et al. 2018; Hunt et al. 2007; Mommen et al. 2014; Pritchard et al. 2015b; Purcell 2004; Purcell and Gorman 2004; Tan et al. 2011). Despite these advances, neoantigens are only rarely detected on cancer cells using mass spectrometry and further studies are required to discover why this is the case (Abelin et al. 2017; Bassani-Sternberg et al. 2016; Carreno et al. 2015; Gloger et al. 2016; Gubin et al. 2014; Hogan et al. 1998; Jarmalavicius et al. 2012; Pritchard et al. 2015b; Yadav et al. 2014).
The majority of recent studies have therefore focused on a forward approach of performing NGS on germline and tumour DNA to identify protein altering mutations that are specific to the cancer cells, followed by epitope prediction via in silico algorithms, as detailed in the following sections.
Prediction of neoantigen(s) in the era of next-generation sequencing technology
The prediction of neoantigens relies on the in silico processing of genomic data and requires knowledge of the donors HLA type, tumour mRNA expression, germline DNA and tumour DNA. The tumour mRNA expression data such as whole genome microarrays (e.g. Pritchard et al. 2015a) or RNA-seq (e.g. Karasaki et al. 2017; van Rooij et al. 2013) are overlaid on tumour-specific cancer mutation information, to identify variants in transcribed genes. These variants are then run through epitope prediction algorithms, to identify peptide sequences that potentially bind to individual-specific HLA-alleles, which requires the amino acid sequence to be translated from the surrounding genetic sequence. A confounding factor is population polymorphism, which if in phase with the somatic variant may additionally alter amino acids from the reference sequence. There are many epitope prediction algorithms available, including SYFPEITHI (Schuler et al. 2007), RANKPEP (Reche et al. 2002), NetMHCpan (Jurtz et al. 2017), NetMHCcons (Karosiene et al. 2012), PickPocket (Zhang et al. 2009), MHCflurry (in pre-print, https://doi.org/10.1101/174243), ANN (Singh and Mishra 2008) and SMM (Peters and Sette 2005). These algorithms employ different prediction models but have all been trained using characterised epitope/MHC combinations, resulting in the prediction of the likelihood of short peptide sequences binding to a given HLA-allele. Bioinformatic pipelines have been created that use whole genome/exome sequencing data and integrate the analysis to include HLA-allele typing, mRNA expression data, peptide processing prediction and HLA-allele binding for the wildtype and mutated peptide. These include pVAC-seq (Hundal et al. 2016), MuPeXi (Bjerregaard et al. 2017a), Cloudneo (Bais et al. 2017) and TIminer (Tappeiner et al. 2017). Immune evasion and editing can be a limitation to specific T-cell immunotherapy, resulting either in failure to initiate tumour clearance or acquired resistance to therapy. This includes loss of HLA expression by chromosomal 6 loss of heterozygosity (LOH) or the down-regulation of support molecules by various methods (e.g. Anagnostou et al. 2017; Chowell et al. 2018; Schrors et al. 2017). To partially address this issue, the computational tool LOHHLA (loss of heterozygosity in human leukocyte antigen) allows allele-specific copy number estimation of the HLA locus from next-generation sequencing data (McGranahan et al. 2017). The refinement of neoantigen prediction by a combination of the above methods will improve the likelihood of immunogenic neoantigens being identified, which has the potential to improve immunotherapeutic approaches targeting neoantigens.
Prediction of MHC-class I compared to MHC-class II neoantigen binding
As the majority of studies identifying peptides that bind to MHC molecules have focused on those recognised by cytotoxic CD8+ T-cells, the prediction of antigen binding to MHC-class I molecules is the most studied. This is influenced both by the function of the cytotoxic CD8+ T-cells in directly triggering programmed cell death and the way in which the peptide fits into the MHC-class I binding groove, which makes the prediction of this binding more amenable to machine learning. Specifically, as previously described, the lack of definitive N- and C-anchor points within the MHC-class II binding groove makes the prediction of peptides that may bind more difficult (Wang et al. 2008).
While the role of the TH1 subset of CD4+ T-cells in priming, supporting, recruiting and proliferation of CD8+ T-cells is well established, CD4+ T-cells recognising immunoreactive MHC-class II-restricted neoantigens have also been described (e.g. Linnemann et al. 2015; Pieper et al. 1999; Tran et al. 2015; Veatch et al. 2018; Wang et al. 1999; reviewed in Sun et al. 2017). The number of MHC-class II-restricted epitopes catalogued is substantially lower compared to MHC-class I. The number of unique peptides of different lengths that are classified as binding to MHC-class I (n = 229,035) and MHC-class II (n = 54,606) in the IEDB database (Vita et al. 2010) https://www.iedb.org/ is shown in Fig. 1, with the most frequently identified peptide length found to bind to MHC-class I being 9 amino acids long and 15 amino acids long for MHC-class II.
MHC-class I peptide consensus binding sequences
In MHC-class I, the population polymorphisms that dictate HLA-subtype can affect the peptide binding groove, including the anchor residues at the N- and C-terminals, resulting in different peptide epitopes preferentially binding depending on amino acid sequence. Once sufficient peptides binding to different HLA subtypes have been characterised, consistent motifs within these amino acid sequences can be identified. Figure 2 shows the consensus motif for six HLA-A subtypes and Fig. 3 shows the motifs for eight HLA-B subtypes. The peptides used to create these motifs were selected from the IEDB, based on those with a sufficient number of peptides to assess (n ≥ 50) and targeting those that have been shown by functional experiment to elicit a T-cell response. These figures illustrate that there are clear anchor motifs at the C- (HLA-A, Fig. 2) and N- (HLA-B, Fig. 3) terminals for the selected HLA subtypes. Additionally, for the more common HLA types (such as HLA-A*01:01 and HLA-A*02:01, Fig. 2 and HLA-B*07:02 and HLA-B*40:02, Fig. 3), consensus binding amino acids are emerging at the N- and C-terminals, respectively. The simple (single amino acid binding) and complex (e.g. common properties of several amino acids) types of common features visible in the consensus logo motifs in Figs. 2 and 3 inform the epitope binding prediction algorithms described in “Types of neoantigen” section.
Consensus logos motifs for peptides binding to selected MHC-class I HLA-A alleles. The logo motifs were created using WebLogo (http://www.weblogo.berkeley.edu/logo.cgi). Peptides for each HLA type were grouped by peptide length and input to WebLogo, with default settings polar amino acids (G, S, T, Y, C, Q, N) are green, basic (K, R, H) blue, acidic (D, E) red and hydrophobic (A, V, L, I, P, W, F, M) amino acids are black. Where sufficient peptides are present (n > 10) for each peptide length, a consensus logo motif was created. a HLA-A*01:01 (n = 147); b HLA-A*02:01 (n = 2536); c HLA-A*03:01 (n = 213); d HLA-A*11:01 (n = 222); e HLA-A*24:02 (n = 282) f HLA-A*68:01 and HLA*68:02 (combined n = 51). (Color figure online)
Consensus logos motifs for peptides binding to selected MHC-class I HLA-B alleles. The logo motifs were created using WebLogo (http://www.weblogo.berkeley.edu/logo.cgi). The overall height of the stack indicates the sequence conservation at that position, while the height of symbols within the stack indicates the relative frequency of each amino or nucleic acid at that position. Peptides for each HLA type were grouped by peptide length and input to WebLogo, with default settings polar amino acids (G, S, T, Y, C, Q, N) are green, basic (K, R, H) blue, acidic (D, E) red and hydrophobic (A, V, L, I, P, W, F, M) amino acids are black. Where sufficient peptides are present (n > 50) for each peptide length, a consensus logo motif was created. a HLA-B*07:02 (n = 337); b HLA-B*15:01, B*15:02, B*15:03 (combined n = 224); c HLA-B*27:01, B*27:02, B*27:04, B*27:05 (combined n = 114); d HLA-B*35:01, B*35:02 (combined n = 296); e HLA-B*40:01, B*40:02 (combined n = 94); f HLA-B*44:01, B*44:02, B*44:03 (combined n = 118); g HLA-B*51:01 (n = 100); h HLA-B*57:01, B*57:03 (combined n = 80). (Color figure online)
Neoantigen-specific T-cell receptors
Antigen-specific TCR gene transfer via patient-derived T-cells provides the opportunity to break self-tolerance and improve the affinity by which cognate TCR binding to ‘self’-derived epitopes can occur. TCR gene therapy has been a successful means of targeting CT and TA antigens (e.g. Morgan et al. 2006; Robbins et al. 2011); however, off target reactivity has been an issue for these therapies, particularly if cross-reactivity between the modified TCR and other ‘self’ antigen occurs (e.g. Cameron et al. 2013; Tan et al. 2015).
As neoantigens occur only within tumour tissue, they do not induce central tolerance and neoantigen-specific TCRs may therefore be more specific and have a higher affinity than TCR targeted to non-mutated antigens. Preclinical studies have shown that the adoptive transfer of neoantigen-specific TCR engineered T-cells can be effective against solid tumours (Bendle et al. 2010; Boulter et al. 2003); however, in practice, unexpected cross-reactivities have hampered their use (e.g. Morgan et al. 2010, 2013), highlighting the need for careful manipulation of the immune system. NGS technologies are enabling comprehensive description of patient-specific TCR repertoires, allowing the identification of the frequencies of unique TCR clonotypes among TILs (reviewed in Rosati et al. 2017). Multiple technologies have emerged for the isolation of TCR genes, allowing the rapid identification of large TCR libraries from intratumoural T-cells. This has therefore facilitated the careful assessment of antigen specificity of intratumoral TCRs independent from primary material. Furthermore, the availability of such TCR gene libraries may facilitate efforts to locate target epitopes within the cancer anti-genome (Hanson et al. 2016; Howie et al. 2015; Kato et al. 2018; Kwong et al. 2009).
Accuracy of epitope prediction in identifying immunoreactive antigens
As each of the algorithms described in the “Types of neoantigen” section are influenced by the training data set, the less common HLA-alleles tend to have less peptides present, resulting in less confident (higher) binding scores. This means these less common HLA subtypes tend to be consistently predicted to bind less well, compared to the more common HLA subtypes (e.g. as assessed in Pritchard et al. 2015a, b). These analyses are extended in Fig. 4, using the epitope dataset stored in the IEDB with proven T-cell stimulating ability (total n = 3632), which were run through four prediction algorithms present in IEDB (ANN, SMM, NetMHCcons and PickPocket). The cumulative percentage plots illustrate that all four prediction algorithms have a similar prediction profile for the most common HLA-A*02 allele; however, the other five HLA-A alleles examined show different abilities of the algorithms to predict similar results. Additionally, for the most common HLA-A*02 allele, just under 50% of the binding scores are < 50 nM, traditionally defined as ‘strong’ binders, while between 70 and 80% of the scores are < 500 nM, traditionally defined as ‘weak’ binders. This pattern is similar for the other HLA-A alleles assessed (HLA-A*01, HLA-A*03, HLA-A*11, HLA-A*24 and HLA-A*68) for ANN and NetMHCcons; however, SMM and PickPocket are more conservative predictors for these less common alleles, with between 0 and 50% of all peptides classified as ‘weak’ binders.
Cumulative percentage plots of epitope prediction score for HLA-A subtypes. Using the epitope dataset present in the IEDB with proven T-cell stimulating ability and known HLA-A subtype (total n = 3451), four prediction algorithms present in IEDB (ANN, SMM, NetMHCcons and PickPocket) were used to provide epitope prediction scores. The cumulative percentage plots were created to show the proportion of scores that fit within the traditional binding scores for ‘strong’ (< 50 nM) and ‘weak’ (< 500 nM) interactions. These values are indicated by the red dotted lines. a HLA-A*01:01 (n = 147); b HLA-A*02:01 (n = 2536); c HLA-A*03:01 (n = 213); d HLA-A*11:01 (n = 222); e HLA-A*24 (n = 282); f HLA-A*68 (combined n = 51)
A further analysis using the IEDB MHC-class I T-cell activating epitopes that bind to both HLA-A and HLA-B alleles (n = 5510) has been carried out. These were run through the same four prediction algorithms as above and then all binding scores graphed (Fig. 5). These graphs illustrate that using the aforementioned standard thresholds to indicate “strong” and “weak” binding, as indicated by the red dotted lines, is likely to miss a large number of epitopes that have been proven to elicit T-cell responses; this is particularly striking in the less common HLA subtypes. These data also further illustrate the differences in binding prediction scores between the different algorithms assessed. Given that these analyses were carried out using functionally characterised immunogenic peptides, an interpretation of these data might be that those algorithms with a large number of prediction scores > 1000 nM are less robust predictors for those HLA-alleles.
Individual peptide scores plotted for each prediction algorithm. Using the IEDB MHC-class I T-cell activating epitopes that bind to defined HLA-A or HLA-B alleles (n = 4814), four prediction algorithms present in IEDB (ANN, SMM, NetMHCcons and PickPocket) were used to provide epitope prediction scores. These are individually plotted to visualise the number of available peptides to assess and the performance of each algorithm against each other and the traditional binding scores for ‘strong’ (< 50 nM) and ‘weak’ (< 500 nM) interactions. These values are indicated by the red dotted lines. (Color figure online)
Data from repositories such as IEDB analysed in a manner such as performed here can be used to aid the selection of the most informative algorithms for the HLA subtypes of interest. Indeed, it has been shown that by analysing immunogenic neopeptides and peptides from the same studies that do not elicit a T-cell response, prediction of binding patterns may be improved (Bjerregaard et al. 2017b). Further, as more studies identify and functionally examine the peptides binding to HLA-alleles are carried out, the more robust these analyses will become; the current interest in this field is significantly increasing available data.
Use of neoantigen prediction in practice
A large number of published studies have assessed the ability of protein altering mutations in cancer cells to form neoantigens using in silico tools (e.g. Balachandran et al. 2017; Brown et al. 2014; Gros et al. 2016; Hodges et al. 2017; Lauss et al. 2017; Linnemann et al. 2015; Luksza et al. 2017; Matsushita et al. 2012; McGranahan et al. 2016; Ock et al. 2017; Pritchard et al. 2015a; Rizvi et al. 2015; Robbins et al. 2013; Snyder et al. 2014; Tran et al. 2016; Van Allen et al. 2015; van Rooij et al. 2013; Verdegaal et al. 2016). While these various tools have an important role to play in the prediction of immunogenic antigens, the majority of the identified epitopes do not initiate an immune response (e.g. as assessed in Gros et al. 2016; Linnemann et al. 2015; McGranahan et al. 2016; Pritchard et al. 2015a; Robbins et al. 2013; Schmidt et al. 2017; Snyder et al. 2014; van Rooij et al. 2013). The possible reasons behind suboptimal immunogenicity of peptide vaccines for cancer are reviewed by Kumai et al. (2017).
A recent study specifically tackled the question of how many predicted epitopes are capable of eliciting an immune response, using the CEF peptide pool (consisting of 32 individual peptides from Cytomegalo-, Epstein–Barr-, and Influenza viruses) and a panel of 42 HLA typed HLA-A*02:01 positive individuals. There were 241 different peptide stimulating CD8+ responses predicted, on the basis of individual HLA-typing. Broadly, 51% of these predictions stimulated a response, of varying strengths, with only 15% occurring in the high frequency range (> 100 spots/400,000 PBMC) and 17% in mid-frequency (> 10 spots/400,000 PBMC) and 19% at the detection limit (1 spot/400,000 PBMC). Fifty-seven unpredicted responses were seen, meaning that of all the responses detected, 68% were predicted and 32% were not. In total, 49% of the predicted peptides were not detectably targeted by CD8+ cells (Moldovan et al. 2016). These data are intriguing on several levels. The first is that the CEF peptides are among the most studied immune responses and that both predicted and unpredicted CD8+ T-cell response after exposure to these peptides are observed indicates there are still significant aspects of epitope prediction that are not yet achieved. The second is that despite prediction in these well characterised antigens, a large proportion did not elicit any detectable immune response. Finally, despite detecting a CD8+ T-cell response to approximately half of the epitopes predicted, only a fraction were a dominant response. Extrapolation of these data to that observed for neoantigen prediction in studies that do not test the potential epitopes for immune cell recognition should cause a pause for thought in how these data are reported and presented.
Testing of neoantigen immunogenicity
It is clear from the published studies that while the data from the prediction algorithms can be used to inform on the potential epitopes created, there still needs to be laboratory testing for immunogenicity of these epitopes. There are a number of methods by which this can be performed, including screening of the predicted peptides across mixed lymphocyte-tumour culture (MLTC) (e.g. Lennerz et al. 2005; Pritchard et al. 2015a), exposure of tandem mini-genes (e.g. Gros et al. 2016; Lu et al. 2014; Mennonna et al. 2017; Tran et al. 2014, 2015) or pMHC multimers (e.g. Cohen et al. 2015; Stronen et al. 2016; van Rooij et al. 2013) to immune cells, and the pulsing of putative peptides with antigen presentation cells (such as dendritic cells or B-cells) and co-culture with T-cells, followed by T-cell exposure to predicted peptide pools (e.g. Rajasagi et al. 2014). These approaches can identify existing memory T-cell immune responses in patients, or reactive naïve T-cells in patients/donors, both of which have potential clinical utility.
Immunotherapeutic potential of neoantigens
Neoantigens have been shown to contribute to the success of various immunotherapies, including the checkpoint inhibitors targeting PD-1/PD-L1/CTLA4 (e.g. Balachandran et al. 2017; Snyder et al. 2014; van Rooij et al. 2013) and other forms of immunotherapy, including dendritic cell vaccines [e.g. unpublished observations and Pritchard et al. 2015a, assessing patients from clinical trials (O’Rourke et al. 2003, 2007)] and adoptive T-cell transfer (e.g. Tran et al. 2016; Verdegaal et al. 2016). Additionally, as neoantigens are capable of stimulating tumour clearance (e.g. Zacharakis et al. 2018), there are currently a number of registered clinical trials that include combination of immunotherapies with a personalised neoantigen component (e.g. with anti-PD-1 checkpoint inhibition NCT02950766, renal cell carcinoma; NCT03199040, triple-negative breast cancer) and dendritic cell vaccine raised against defined neoantigens (e.g. NCT03300843, melanoma, gastrointestinal, breast, ovarian and pancreatic cancers, NCT03558945, pancreatic cancer) and adjuvant personalised neoantigen peptide vaccine, with the immunostimulant poly-ICLC (e.g. NCT02510950, glioblastoma and astrocytoma and NCT01970358, melanoma). Although it may be possible to target the more common ‘driver’ gene mutations such as BRAF (Sharkey et al. 2004; Somasundaram et al. 2006; Veatch et al. 2018), KRAS (Bergmann-Leitner et al. 1998; Shono et al. 2003), p53 (Ichiki et al. 2004) and NRAS (Linard et al. 2002), the required combination of non-synonymous variant and specific HLA-allele make neoantigens more likely to be a personalised therapy option.
As illustrated with examples above, the major limitation to the use of neoantigens in immunotherapy is the reliable personalised prediction of those that will have undergone proteasomal cleavage, transport to the ER, binding to the individuals HLA molecule and recognition by the T-cell receptor to stimulate an immune response capable of tumour clearance.
Conclusion
The somatic mutations acquired by cancer cells can be recognised as ‘non-self’ by the immune system and are capable of inducing an immune response that can selectively target and remove tumour cells. There are a number of steps required in order for the peptides to be displayed to the immune system and each of these processes has optimal conditions under which they occur. Therefore, despite there being a large number of potential neoantigens in some cancers with high mutation burden, only a fraction are able to ultimately mount an immune response. With the improvement in molecular and in silico capabilities in recent years, the number of identified immunogenic neoantigens has substantially increased. As demonstrated here, the current methods do not consistently identify epitopes that are a priori known to mount an immune response. With a greater number of verified neoantigens, the better the ability of trained in silico prediction tools to reliably identify those that may have clinical utility. There is considerable potential in the use of neoantigens to treat patients, either alone or in combination with other immunotherapies and with continued advancements, these potentials will be realised.
References
Abelin JG, Keskin DB, Sarkizova S, Hartigan CR, Zhang W, Sidney J, Stevens J, Lane W, Zhang GL, Eisenhaure TM, Clauser KR, Hacohen N, Rooney MS, Carr SA, Wu CJ (2017) Mass spectrometry profiling of HLA-associated peptidomes in mono-allelic cells enables more accurate epitope prediction. Immunity 46:315–326
Akcakanat A, Kanda T, Koyama Y, Watanabe M, Kimura E, Yoshida Y, Komukai S, Nakagawa S, Odani S, Fujii H, Hatakeyama K (2004) NY-ESO-1 expression and its serum immunoreactivity in esophageal cancer. Cancer Chemother Pharmacol 54:95–100
Akers SN, Odunsi K, Karpf AR (2010) Regulation of cancer germline antigen gene expression: implications for cancer immunotherapy. Future Oncol 6:717–732
Aleksic M, Liddy N, Molloy PE, Pumphrey N, Vuidepot A, Chang KM, Jakobsen BK (2012) Different affinity windows for virus and cancer-specific T-cell receptors: implications for therapeutic strategies. Eur J Immunol 42:3174–3179
Anagnostou V, Smith KN, Forde PM, Niknafs N, Bhattacharya R, White J, Zhang T, Adleff V, Phallen J, Wali N, Hruban C, Guthrie VB, Rodgers K, Naidoo J, Kang H, Sharfman W, Georgiades C, Verde F, Illei P, Li QK, Gabrielson E, Brock MV, Zahnow CA, Baylin SB, Scharpf RB, Brahmer JR, Karchin R, Pardoll DM, Velculescu VE (2017) Evolution of neoantigen landscape during immune checkpoint blockade in non-small cell lung cancer. Cancer Discov 7:264–276
Ayyoub M, Rimoldi D, Guillaume P, Romero P, Cerottini JC, Valmori D, Speiser D (2003) Tumor-reactive, SSX-2-specific CD8 + T cells are selectively expanded during immune responses to antigen-expressing tumors in melanoma patients. Cancer Res 63:5601–5606
Bais P, Namburi S, Gatti DM, Zhang X, Chuang JH (2017) CloudNeo: a cloud pipeline for identifying patient-specific tumor neoantigens. Bioinformatics 33:3110–3112
Balachandran VP, Luksza M, Zhao JN, Makarov V, Moral JA, Remark R, Herbst B, Askan G, Bhanot U, Senbabaoglu Y, Wells DK, Cary CIO, Grbovic-Huezo O, Attiyeh M, Medina B, Zhang J, Loo J, Saglimbeni J, Abu-Akeel M, Zappasodi R, Riaz N, Smoragiewicz M, Kelley ZL, Basturk O, Australian Pancreatic Cancer Genome I, Garvan Institute of Medical R, Prince of Wales H, Royal North Shore H, University of G, St Vincent’s H, Institute QBMR, University of Melbourne CfCR, University of Queensland IfMB, Bankstown H, Liverpool H, Royal Prince Alfred Hospital COBL, Westmead H, Fremantle H, St John of God H, Royal Adelaide H, Flinders Medical C, Envoi P, Princess Alexandria H, Austin H, Johns Hopkins Medical I, Cancer AR-NCfARo, Gonen M, Allen AJ, Fearon PJ, Merad DT, Gnjatic M, Iacobuzio-Donahue CA, Wolchok JD, DeMatteo RP, Chan TA, Greenbaum BD, Merghoub T, Leach SD (2017). Identification of unique neoantigen qualities in long-term survivors of pancreatic cancer. Nature 551:512–516
Barfoed AM, Petersen TR, Kirkin AF, Thor Straten P, Claesson MH, Zeuthen J (2000) Cytotoxic T-lymphocyte clones, established by stimulation with the HLA-A2 binding p5365-73 wild type peptide loaded on dendritic cells In vitro, specifically recognize and lyse HLA-A2 tumour cells overexpressing the p53 protein. Scand J Immunol 51:128–133
Basler M, Kirk CJ, Groettrup M (2013) The immunoproteasome in antigen processing and other immunological functions. Curr Opin Immunol 25:74–80
Bassani-Sternberg M, Braunlein E, Klar R, Engleitner T, Sinitcyn P, Audehm S, Straub M, Weber J, Slotta-Huspenina J, Specht K, Martignoni ME, Werner A, Hein R, D HB, Peschel C, Rad R, Cox J, Mann M, Krackhardt AM (2016) Direct identification of clinically relevant neoepitopes presented on native human melanoma tissue by mass spectrometry. Nat Commun 7:13404
Baylin SB, Jones PA (2011) A decade of exploring the cancer epigenome—biological and translational implications. Nat Rev Cancer 11:726–734
Bendle GM, Linnemann C, Hooijkaas AI, Bies L, de Witte MA, Jorritsma A, Kaiser AD, Pouw N, Debets R, Kieback E, Uckert W, Song JY, Haanen JB, Schumacher TN (2010) Lethal graft-versus-host disease in mouse models of T cell receptor gene therapy. Nat Med 16:565–570 (561p following 570)
Bentzen AK, Marquard AM, Lyngaa R, Saini SK, Ramskov S, Donia M, Such L, Furness AJ, McGranahan N, Rosenthal R, Straten PT, Szallasi Z, Svane IM, Swanton C, Quezada SA, Jakobsen SN, Eklund AC, Hadrup SR (2016) Large-scale detection of antigen-specific T cells using peptide-MHC-I multimers labeled with DNA barcodes. Nat Biotechnol 34:1037–1045
Bergmann-Leitner ES, Kantor JA, Shupert WL, Schlom J, Abrams SI (1998) Identification of a human CD8 + T lymphocyte neo-epitope created by a ras codon 12 mutation which is restricted by the HLA-A2 allele. Cell Immunol 187:103–116
Bhasin M, Lata S, Raghava GP (2007) TAPPred prediction of TAP-binding peptides in antigens. Methods Mol Biol 409:381–386
Bjerregaard AM, Nielsen M, Hadrup SR, Szallasi Z, Eklund AC (2017a) MuPeXI: prediction of neo-epitopes from tumor sequencing data. Cancer Immunol Immunother 66:1123–1130
Bjerregaard AM, Nielsen M, Jurtz V, Barra CM, Hadrup SR, Szallasi Z, Eklund AC (2017b) An analysis of natural T cell responses to predicted tumor neoepitopes. Front Immunol 8:1566
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC (1987a) The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature 329:512–518
Bjorkman PJ, Saper MA, Samraoui B, Bennett WS, Strominger JL, Wiley DC (1987b) Structure of the human class I histocompatibility antigen, HLA-A2. Nature 329:506–512
Blankenstein T, Leisegang M, Uckert W, Schreiber H (2015) Targeting cancer-specific mutations by T cell receptor gene therapy. Curr Opin Immunol 33:112–119
Blum JS, Wearsch PA, Cresswell P (2013) Pathways of antigen processing. Annu Rev Immunol 31:443–473
Bollard CM, Gottschalk S, Torrano V, Diouf O, Ku S, Hazrat Y, Carrum G, Ramos C, Fayad L, Shpall EJ, Pro B, Liu H, Wu MF, Lee D, Sheehan AM, Zu Y, Gee AP, Brenner MK, Heslop HE, Rooney CM (2014) Sustained complete responses in patients with lymphoma receiving autologous cytotoxic T lymphocytes targeting Epstein-Barr virus latent membrane proteins. J Clin Oncol 32:798–808
Boulter JM, Glick M, Todorov PT, Baston E, Sami M, Rizkallah P, Jakobsen BK (2003) Stable, soluble T-cell receptor molecules for crystallization and therapeutics. Protein Eng 16:707–711
Brichard V, Van Pel A, Wolfel T, Wolfel C, De Plaen E, Lethe B, Coulie P, Boon T (1993) The tyrosinase gene codes for an antigen recognized by autologous cytolytic T lymphocytes on HLA-A2 melanomas. J Exp Med 178:489–495
Brown SD, Warren RL, Gibb EA, Martin SD, Spinelli JJ, Nelson BH, Holt RA (2014) Neo-antigens predicted by tumor genome meta-analysis correlate with increased patient survival. Genome Res 24:743–750
Calis JJ, Maybeno M, Greenbaum JA, Weiskopf D, De Silva AD, Sette A, Kesmir C, Peters B (2013) Properties of MHC class I presented peptides that enhance immunogenicity. PLoS Comput Biol 9:e1003266
Cameron BJ, Gerry AB, Dukes J, Harper JV, Kannan V, Bianchi FC, Grand F, Brewer JE, Gupta M, Plesa G, Bossi G, Vuidepot A, Powlesland AS, Legg A, Adams KJ, Bennett AD, Pumphrey NJ, Williams DD, Binder-Scholl G, Kulikovskaya I, Levine BL, Riley JL, Varela-Rohena A, Stadtmauer EA, Rapoport AP, Linette GP, June CH, Hassan NJ, Kalos M, Jakobsen BK (2013) Identification of a Titin-derived HLA-A1-presented peptide as a cross-reactive target for engineered MAGE A3-directed T cells. Sci Transl Med 5:197ra103
Carreno BM, Magrini V, Becker-Hapak M, Kaabinejadian S, Hundal J, Petti AA, Ly A, Lie WR, Hildebrand WH, Mardis ER, Linette GP (2015) Cancer immunotherapy. A dendritic cell vaccine increases the breadth and diversity of melanoma neoantigen-specific T cells. Science 348:803–808
Chang TC, Carter RA, Li Y, Li Y, Wang H, Edmonson MN, Chen X, Arnold P, Geiger TL, Wu G, Peng J, Dyer M, Downing JR, Green DR, Thomas PG, Zhang J (2017) The neoepitope landscape in pediatric cancers. Genome Med 9:78
Chen YT, Hsu M, Lee P, Shin SJ, Mhawech-Fauceglia P, Odunsi K, Altorki NK, Song CJ, Jin BQ, Simpson AJ, Old LJ (2009) Cancer/testis antigen CT45: analysis of mRNA and protein expression in human cancer. Int J Cancer 124:2893–2898
Chomez P, De Backer O, Bertrand M, De Plaen E, Boon T, Lucas S (2001) An overview of the MAGE gene family with the identification of all human members of the family. Cancer Res 61:5544–5551
Chowell D, Morris LGT, Grigg CM, Weber JK, Samstein RM, Makarov V, Kuo F, Kendall SM, Requena D, Riaz N, Greenbaum B, Carroll J, Garon E, Hyman DM, Zehir A, Solit D, Berger M, Zhou R, Rizvi NA, Chan TA (2018) Patient HLA class I genotype influences cancer response to checkpoint blockade immunotherapy. Science 359:582–587
Cloosen S, Arnold J, Thio M, Bos GM, Kyewski B, Germeraad WT (2007) Expression of tumor-associated differentiation antigens, MUC1 glycoforms and CEA, in human thymic epithelial cells: implications for self-tolerance and tumor therapy. Cancer Res 67:3919–3926
Cohen CJ, Gartner JJ, Horovitz-Fried M, Shamalov K, Trebska-McGowan K, Bliskovsky VV, Parkhurst MR, Ankri C, Prickett TD, Crystal JS, Li YF, El-Gamil M, Rosenberg SA, Robbins PF (2015) Isolation of neoantigen-specific T cells from tumor and peripheral lymphocytes. J Clin Invest 125:3981–3991
Coulie PG, Van den Eynde BJ, van der Bruggen P, Boon T (2014) Tumour antigens recognized by T lymphocytes: at the core of cancer immunotherapy. Nat Rev Cancer 14:135–146
De Plaen E, Lurquin C, Van Pel A, Mariame B, Szikora JP, Wolfel T, Sibille C, Chomez P, Boon T (1988) Immunogenic (tum-) variants of mouse tumor P815: cloning of the gene of tum- antigen P91A and identification of the tum-mutation. Proc Natl Acad Sci USA 85:2274–2278
De Smet C, De Backer O, Faraoni I, Lurquin C, Brasseur F, Boon T (1996) The activation of human gene MAGE-1 in tumor cells is correlated with genome-wide demethylation. Proc Natl Acad Sci USA 93:7149–7153
De Smet C, Lurquin C, Lethe B, Martelange V, Boon T (1999) DNA methylation is the primary silencing mechanism for a set of germ line- and tumor-specific genes with a CpG-rich promoter. Mol Cell Biol 19:7327–7335
dos Santos NR, Torensma R, de Vries TJ, Schreurs MW, de Bruijn DR, Kater-Baats E, Ruiter DJ, Adema GJ, van Muijen GN, van Kessel AG (2000) Heterogeneous expression of the SSX cancer/testis antigens in human melanoma lesions and cell lines. Cancer Res 60:1654–1662
Ellsworth RE, Ellsworth DL, Patney HL, Deyarmin B, Love B, Hooke JA, Shriver CD (2008) Amplification of HER2 is a marker for global genomic instability. BMC Cancer 8:297
Emmerich NP, Nussbaum AK, Stevanovic S, Priemer M, Toes RE, Rammensee HG, Schild H (2000) The human 26 S and 20 S proteasomes generate overlapping but different sets of peptide fragments from a model protein substrate. J Biol Chem 275:21140–21148
Fiszer D, Kurpisz M (1998) Major histocompatibility complex expression on human, male germ cells: a review. Am J Reprod Immunol 40:172–176
Fratta E, Coral S, Covre A, Parisi G, Colizzi F, Danielli R, Nicolay HJ, Sigalotti L, Maio M (2011) The biology of cancer testis antigens: putative function, regulation and therapeutic potential. Mol Oncol 5:164–182
Fremont DH, Matsumura M, Stura EA, Peterson PA, Wilson IA (1992) Crystal structures of two viral peptides in complex with murine MHC class I H-2Kb. Science 257:919–927
Freudenmann LK, Marcu A, Stevanovic S (2018) Mapping the tumour human leukocyte antigen (HLA) ligandome by mass spectrometry. Immunology 154:331–345
Gillison ML, Koch WM, Capone RB, Spafford M, Westra WH, Wu L, Zahurak ML, Daniel RW, Viglione M, Symer DE, Shah KV, Sidransky D (2000) Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst 92:709–720
Gloger A, Ritz D, Fugmann T, Neri D (2016) Mass spectrometric analysis of the HLA class I peptidome of melanoma cell lines as a promising tool for the identification of putative tumor-associated HLA epitopes. Cancer Immunol Immunother 65:1377–1393
Gnjatic S, Nishikawa H, Jungbluth AA, Gure AO, Ritter G, Jager E, Knuth A, Chen YT, Old LJ (2006) NY-ESO-1: review of an immunogenic tumor antigen. Adv Cancer Res 95:1–30
Goodwin S, McPherson JD, McCombie WR (2016) Coming of age: ten years of next-generation sequencing technologies. Nat Rev Genet 17:333–351
Grakoui A, Bromley SK, Sumen C, Davis MM, Shaw AS, Allen PM, Dustin ML (1999) The immunological synapse: a molecular machine controlling T cell activation. Science 285:221–227
Greve KB, Pohl M, Olsen KE, Nielsen O, Ditzel HJ, Gjerstorff MF (2014) SSX2-4 expression in early-stage non-small cell lung cancer. Tissue Antigens 83:344–349
Gros A, Parkhurst MR, Tran E, Pasetto A, Robbins PF, Ilyas S, Prickett TD, Gartner JJ, Crystal JS, Roberts IM, Trebska-McGowan K, Wunderlich JR, Yang JC, Rosenberg SA (2016) Prospective identification of neoantigen-specific lymphocytes in the peripheral blood of melanoma patients. Nat Med 22:433–438
Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, Mulder GE, Toebes M, Vesely MD, Lam SS, Korman AJ, Allison JP, Freeman GJ, Sharpe AH, Pearce EL, Schumacher TN, Aebersold R, Rammensee HG, Melief CJ, Mardis ER, Gillanders WE, Artyomov MN, Schreiber RD (2014) Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature 515:577–581
Guo HC, Jardetzky TS, Garrett TP, Lane WS, Strominger JL, Wiley DC (1992) Different length peptides bind to HLA-Aw68 similarly at their ends but bulge out in the middle. Nature 360:364–366
Hanson WM, Chen Z, Jackson LK, Attaf M, Sewell AK, Heemstra JM, Phillips JD (2016) Reversible oligonucleotide chain blocking enables bead capture and amplification of T-cell receptor alpha and beta chain mRNAs. J Am Chem Soc 138:11073–11076
Hayward NK, Wilmott JS, Waddell N, Johansson PA, Field MA, Nones K, Patch AM, Kakavand H, Alexandrov LB, Burke H, Jakrot V, Kazakoff S, Holmes O, Leonard C, Sabarinathan R, Mularoni L, Wood S, Xu Q, Waddell N, Tembe V, Pupo GM, De Paoli-Iseppi R, Vilain RE, Shang P, Lau LMS, Dagg RA, Schramm SJ, Pritchard A, Dutton-Regester K, Newell F, Fitzgerald A, Shang CA, Grimmond SM, Pickett HA, Yang JY, Stretch JR, Behren A, Kefford RF, Hersey P, Long GV, Cebon J, Shackleton M, Spillane AJ, Saw RPM, Lopez-Bigas N, Pearson JV, Thompson JF, Scolyer RA, Mann GJ (2017) Whole-genome landscapes of major melanoma subtypes. Nature 545:175–180
Heslop HE, Slobod KS, Pule MA, Hale GA, Rousseau A, Smith CA, Bollard CM, Liu H, Wu MF, Rochester RJ, Amrolia PJ, Hurwitz JL, Brenner MK, Rooney CM (2010) Long-term outcome of EBV-specific T-cell infusions to prevent or treat EBV-related lymphoproliferative disease in transplant recipients. Blood 115:925–935
Hodges TR, Ott M, Xiu J, Gatalica Z, Swensen J, Zhou S, Huse JT, de Groot J, Li S, Overwijk WW, Spetzler D, Heimberger AB (2017) Mutational burden, immune checkpoint expression, and mismatch repair in glioma: implications for immune checkpoint immunotherapy. Neuro-oncology 19:1047–1057
Hofmann O, Caballero OL, Stevenson BJ, Chen YT, Cohen T, Chua R, Maher CA, Panji S, Schaefer U, Kruger A, Lehvaslaiho M, Carninci P, Hayashizaki Y, Jongeneel CV, Simpson AJ, Old LJ, Hide W (2008) Genome-wide analysis of cancer/testis gene expression. Proc Natl Acad Sci USA 105:20422–20427
Hogan KT, Eisinger DP, Cupp SB III, Lekstrom KJ, Deacon DD, Shabanowitz J, Hunt DF, Engelhard VH, Slingluff CL Jr, Ross MM (1998) The peptide recognized by HLA-A68.2-restricted, squamous cell carcinoma of the lung-specific cytotoxic T lymphocytes is derived from a mutated elongation factor 2 gene. Cancer Res 58:5144–5150
Howie B, Sherwood AM, Berkebile AD, Berka J, Emerson RO, Williamson DW, Kirsch I, Vignali M, Rieder MJ, Carlson CS, Robins HS (2015) High-throughput pairing of T cell receptor alpha and beta sequences. Sci Transl Med 7:301ra131
Hundal J, Carreno BM, Petti AA, Linette GP, Griffith OL, Mardis ER, Griffith M (2016) pVAC-Seq: a genome-guided in silico approach to identifying tumor neoantigens. Genome Med 8:11
Hunt DF, Henderson RA, Shabanowitz J, Sakaguchi K, Michel H, Sevilir N, Cox AL, Appella E, Engelhard VH (2007) Pillars article: characterization of peptides bound to the class I MHC molecule HLA-A2.1 by mass spectrometry. Science 1992. 255:1261–1263. J Immunol 179:2669–2671
Ichiki Y, Takenoyama M, Mizukami M, So T, Sugaya M, Yasuda M, So T, Hanagiri T, Sugio K, Yasumoto K (2004) Simultaneous cellular and humoral immune response against mutated p53 in a patient with lung cancer. J Immunol 172:4844–4850
Ilyas S, Yang JC (2015) Landscape of tumor antigens in T Cell Immunotherapy. J Immunol 195:5117–5122
Inderberg EM, Walchli S, Myhre MR, Trachsel S, Almasbak H, Kvalheim G, Gaudernack G (2017) T cell therapy targeting a public neoantigen in microsatellite instable colon cancer reduces in vivo tumor growth. Oncoimmunology 6:e1302631
Jager E, Chen YT, Drijfhout JW, Karbach J, Ringhoffer M, Jager D, Arand M, Wada H, Noguchi Y, Stockert E, Old LJ, Knuth A (1998) Simultaneous humoral and cellular immune response against cancer-testis antigen NY-ESO-1: definition of human histocompatibility leukocyte antigen (HLA)-A2-binding peptide epitopes. J Exp Med 187:265–270
Jarmalavicius S, Welte Y, Walden P (2012) High immunogenicity of the human leukocyte antigen peptidomes of melanoma tumor cells. J Biol Chem 287:33401–33411
Johnson LA, Morgan RA, Dudley ME, Cassard L, Yang JC, Hughes MS, Kammula US, Royal RE, Sherry RM, Wunderlich JR, Lee CC, Restifo NP, Schwarz SL, Cogdill AP, Bishop RJ, Kim H, Brewer CC, Rudy SF, VanWaes C, Davis JL, Mathur A, Ripley RT, Nathan DA, Laurencot CM, Rosenberg SA (2009) Gene therapy with human and mouse T-cell receptors mediates cancer regression and targets normal tissues expressing cognate antigen. Blood 114:535–546
Jurtz V, Paul S, Andreatta M, Marcatili P, Peters B, Nielsen M (2017) NetMHCpan-4.0: improved peptide-MHC class I interaction predictions integrating eluted ligand and peptide binding affinity data. J Immunol 199:3360–3368
Kalejs M, Erenpreisa J (2005) Cancer/testis antigens and gametogenesis: a review and “brain-storming” session. Cancer Cell Int 5:4
Karasaki T, Nagayama K, Kuwano H, Nitadori JI, Sato M, Anraku M, Hosoi A, Matsushita H, Takazawa M, Ohara O, Nakajima J, Kakimi K (2017) Prediction and prioritization of neoantigens: integration of RNA sequencing data with whole-exome sequencing. Cancer Sci 108:170–177
Karosiene E, Lundegaard C, Lund O, Nielsen M (2012) NetMHCcons: a consensus method for the major histocompatibility complex class I predictions. Immunogenetics 64:177–186
Karpf AR, Jones DA (2002) Reactivating the expression of methylation silenced genes in human cancer. Oncogene 21:5496–5503
Kato T, Matsuda T, Ikeda Y, Park JH, Leisegang M, Yoshimura S, Hikichi T, Harada M, Zewde M, Sato S, Hasegawa K, Kiyotani K, Nakamura Y (2018) Effective screening of T cells recognizing neoantigens and construction of T-cell receptor-engineered T cells. Oncotarget 9:11009–11019
Kesmir C, Nussbaum AK, Schild H, Detours V, Brunak S (2002) Prediction of proteasome cleavage motifs by neural networks. Protein Eng 15:287–296
Kessler JH, Beekman NJ, Bres-Vloemans SA, Verdijk P, van Veelen PA, Kloosterman-Joosten AM, Vissers DC, ten Bosch GJ, Kester MG, Sijts A, Wouter Drijfhout J, Ossendorp F, Offringa R, Melief CJ (2001) Efficient identification of novel HLA-A(*)0201-presented cytotoxic T lymphocyte epitopes in the widely expressed tumor antigen PRAME by proteasome-mediated digestion analysis. J Exp Med 193:73–88
Kloetzel PM (2004) The proteasome and MHC class I antigen processing. Biochim Biophys Acta 1695:225–233
Knuth A, Danowski B, Oettgen HF, Old LJ (1984) T-cell-mediated cytotoxicity against autologous malignant melanoma: analysis with interleukin 2-dependent T-cell cultures. Proc Natl Acad Sci USA 81:3511–3515
Kumai T, Kobayashi H, Harabuchi Y, Celis E (2017) Peptide vaccines in cancer-old concept revisited. Curr Opin Immunol 45:1–7
Kwong GA, Radu CG, Hwang K, Shu CJ, Ma C, Koya RC, Comin-Anduix B, Hadrup SR, Bailey RC, Witte ON, Schumacher TN, Ribas A, Heath JR (2009) Modular nucleic acid assembled p/MHC microarrays for multiplexed sorting of antigen-specific T cells. J Am Chem Soc 131:9695–9703
Lauss M, Donia M, Harbst K, Andersen R, Mitra S, Rosengren F, Salim M, Vallon-Christersson J, Torngren T, Kvist A, Ringner M, Svane IM, Jonsson G (2017) Mutational and putative neoantigen load predict clinical benefit of adoptive T cell therapy in melanoma. Nat Commun 8:1738
Lawrence MS, Stojanov P, Polak P, Kryukov GV, Cibulskis K, Sivachenko A, Carter SL, Stewart C, Mermel CH, Roberts SA, Kiezun A, Hammerman PS, McKenna A, Drier Y, Zou L, Ramos AH, Pugh TJ, Stransky N, Helman E, Kim J, Sougnez C, Ambrogio L, Nickerson E, Shefler E, Cortes ML, Auclair D, Saksena G, Voet D, Noble M, DiCara D, Lin P, Lichtenstein L, Heiman DI, Fennell T, Imielinski M, Hernandez B, Hodis E, Baca S, Dulak AM, Lohr J, Landau DA, Wu CJ, Melendez-Zajgla J, Hidalgo-Miranda A, Koren A, McCarroll SA, Mora J, Crompton B, Onofrio R, Parkin M, Winckler W, Ardlie K, Gabriel SB, Roberts CWM, Biegel JA, Stegmaier K, Bass AJ, Garraway LA, Meyerson M, Golub TR, Gordenin DA, Sunyaev S, Lander ES, Getz G (2013) Mutational heterogeneity in cancer and the search for new cancer-associated genes. Nature 499:214–218
Lehnert E, Tampe R (2017) Structure and dynamics of antigenic peptides in complex with TAP. Front Immunol 8:10
Lennerz V, Fatho M, Gentilini C, Frye RA, Lifke A, Ferel D, Wolfel C, Huber C, Wolfel T (2005) The response of autologous T cells to a human melanoma is dominated by mutated neoantigens. Proc Natl Acad Sci USA 102:16013–16018
Linard B, Bezieau S, Benlalam H, Labarriere N, Guilloux Y, Diez E, Jotereau F (2002) A ras-mutated peptide targeted by CTL infiltrating a human melanoma lesion. J Immunol 168:4802–4808
Linnebacher M, Gebert J, Rudy W, Woerner S, Yuan YP, Bork P, von Knebel Doeberitz M (2001) Frameshift peptide-derived T-cell epitopes: a source of novel tumor-specific antigens. Int J Cancer 93:6–11
Linnemann C, Mezzadra R, Schumacher TN (2014) TCR repertoires of intratumoral T-cell subsets. Immunol Rev 257:72–82
Linnemann C, van Buuren MM, Bies L, Verdegaal EM, Schotte R, Calis JJ, Behjati S, Velds A, Hilkmann H, Atmioui DE, Visser M, Stratton MR, Haanen JB, Spits H, van der Burg SH, Schumacher TN (2015) High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4 + T cells in human melanoma. Nat Med 21:81–85
Louis CU, Straathof K, Bollard CM, Ennamuri S, Gerken C, Lopez TT, Huls MH, Sheehan A, Wu MF, Liu H, Gee A, Brenner MK, Rooney CM, Heslop HE, Gottschalk S (2010) Adoptive transfer of EBV-specific T cells results in sustained clinical responses in patients with locoregional nasopharyngeal carcinoma. J Immunother 33:983–990
Lu YC, Robbins PF (2016) Cancer immunotherapy targeting neoantigens. Semin Immunol 28:22–27
Lu YC, Yao X, Crystal JS, Li YF, El-Gamil M, Gross C, Davis L, Dudley ME, Yang JC, Samuels Y, Rosenberg SA, Robbins PF (2014) Efficient identification of mutated cancer antigens recognized by T cells associated with durable tumor regressions. Clin Cancer Res 20:3401–3410
Luimstra JJ, Garstka MA, Roex MCJ, Redeker A, Janssen GMC, van Veelen PA, Arens R, Falkenburg JHF, Neefjes J, Ovaa H (2018) A flexible MHC class I multimer loading system for large-scale detection of antigen-specific T cells. J Exp Med 215:1493–1504
Luksza M, Riaz N, Makarov V, Balachandran VP, Hellmann MD, Solovyov A, Rizvi NA, Merghoub T, Levine AJ, Chan TA, Wolchok JD, Greenbaum BD (2017) A neoantigen fitness model predicts tumour response to checkpoint blockade immunotherapy. Nature 551:517–520
Lupetti R, Pisarra P, Verrecchia A, Farina C, Nicolini G, Anichini A, Bordignon C, Sensi M, Parmiani G, Traversari C (1998) Translation of a retained intron in tyrosinase-related protein (TRP) 2 mRNA generates a new cytotoxic T lymphocyte (CTL)-defined and shared human melanoma antigen not expressed in normal cells of the melanocytic lineage. J Exp Med 188:1005–1016
Matsushita H, Vesely MD, Koboldt DC, Rickert CG, Uppaluri R, Magrini VJ, Arthur CD, White JM, Chen YS, Shea LK, Hundal J, Wendl MC, Demeter R, Wylie T, Allison JP, Smyth MJ, Old LJ, Mardis ER, Schreiber RD (2012) Cancer exome analysis reveals a T-cell-dependent mechanism of cancer immunoediting. Nature 482:400–404
McGranahan N, Furness AJ, Rosenthal R, Ramskov S, Lyngaa R, Saini SK, Jamal-Hanjani M, Wilson GA, Birkbak NJ, Hiley CT, Watkins TB, Shafi S, Murugaesu N, Mitter R, Akarca AU, Linares J, Marafioti T, Henry JY, Van Allen EM, Miao D, Schilling B, Schadendorf D, Garraway LA, Makarov V, Rizvi NA, Snyder A, Hellmann MD, Merghoub T, Wolchok JD, Shukla SA, Wu CJ, Peggs KS, Chan TA, Hadrup SR, Quezada SA, Swanton C (2016) Clonal neoantigens elicit T cell immunoreactivity and sensitivity to immune checkpoint blockade. Science 351:1463–1469
McGranahan N, Rosenthal R, Hiley CT, Rowan AJ, Watkins TBK, Wilson GA, Birkbak NJ, Veeriah S, Van Loo P, Herrero J, Swanton C (2017) Allele-specific HLA loss and immune escape in lung cancer evolution. Cell 171:1259–1271.e1211
Mennonna D, Maccalli C, Romano MC, Garavaglia C, Capocefalo F, Bordoni R, Severgnini M, De Bellis G, Sidney J, Sette A, Gori A, Longhi R, Braga M, Ghirardelli L, Baldari L, Orsenigo E, Albarello L, Zino E, Fleischhauer K, Mazzola G, Ferrero N, Amoroso A, Casorati G, Parmiani G, Dellabona P (2017) T cell neoepitope discovery in colorectal cancer by high throughput profiling of somatic mutations in expressed genes. Gut 66:454–463
Milne K, Barnes RO, Girardin A, Mawer MA, Nesslinger NJ, Ng A, Nielsen JS, Sahota R, Tran E, Webb JR, Wong MQ, Wick DA, Wray A, McMurtrie E, Kobel M, Kalloger SE, Gilks CB, Watson PH, Nelson BH (2008) Tumor-infiltrating T cells correlate with NY-ESO-1-specific autoantibodies in ovarian cancer. PLoS ONE 3:e3409
Moldovan I, Targoni O, Zhang W, Sundararaman S, Lehmann PV (2016) How frequently are predicted peptides actually recognized by CD8 cells? Cancer Immunol Immunother 65:847–855
Mommen GP, Frese CK, Meiring HD, van Gaans-van den Brink J, de Jong AP, van Els CA, Heck AJ (2014) Expanding the detectable HLA peptide repertoire using electron-transfer/higher-energy collision dissociation (EThcD). Proc Natl Acad Sci USA 111:4507–4512
Monach PA, Meredith SC, Siegel CT, Schreiber H (1995) A unique tumor antigen produced by a single amino acid substitution. Immunity 2:45–59
Monks CR, Freiberg BA, Kupfer H, Sciaky N, Kupfer A (1998) Three-dimensional segregation of supramolecular activation clusters in T cells. Nature 395:82–86
Morgan RA, Dudley ME, Wunderlich JR, Hughes MS, Yang JC, Sherry RM, Royal RE, Topalian SL, Kammula US, Restifo NP, Zheng Z, Nahvi A, de Vries CR, Rogers-Freezer LJ, Mavroukakis SA, Rosenberg SA (2006) Cancer regression in patients after transfer of genetically engineered lymphocytes. Science 314:126–129
Morgan RA, Yang JC, Kitano M, Dudley ME, Laurencot CM, Rosenberg SA (2010) Case report of a serious adverse event following the administration of T cells transduced with a chimeric antigen receptor recognizing ERBB2. Mol Ther 18:843–851
Morgan RA, Chinnasamy N, Abate-Daga D, Gros A, Robbins PF, Zheng Z, Dudley ME, Feldman SA, Yang JC, Sherry RM, Phan GQ, Hughes MS, Kammula US, Miller AD, Hessman CJ, Stewart AA, Restifo NP, Quezado MM, Alimchandani M, Rosenberg AZ, Nath A, Wang T, Bielekova B, Wuest SC, Akula N, McMahon FJ, Wilde S, Mosetter B, Schendel DJ, Laurencot CM, Rosenberg SA (2013) Cancer regression and neurological toxicity following anti-MAGE-A3 TCR gene therapy. J Immunother 36:133–151
Murray RJ, Kurilla MG, Brooks JM, Thomas WA, Rowe M, Kieff E, Rickinson AB (1992) Identification of target antigens for the human cytotoxic T cell response to Epstein-Barr virus (EBV): implications for the immune control of EBV-positive malignancies. J Exp Med 176:157–168
Natarajan K, Li H, Mariuzza RA, Margulies DH (1999) MHC class I molecules, structure and function. Rev Immunogenet 1:32–46
Neller MA, Lopez JA, Schmidt CW (2008) Antigens for cancer immunotherapy. Semin Immunol 20:286–295
Nelson CA, Fremont DH (1999) Structural principles of MHC class II antigen presentation. Rev Immunogenet 1:47–59
O’Rourke MG, Johnson M, Lanagan C, See J, Yang J, Bell JR, Slater GJ, Kerr BM, Crowe B, Purdie DM, Elliott SL, Ellem KA, Schmidt CW (2003) Durable complete clinical responses in a phase I/II trial using an autologous melanoma cell/dendritic cell vaccine. Cancer Immunol Immunother 52:387–395
O’Rourke MG, Johnson MK, Lanagan CM, See JL, O’Connor LE, Slater GJ, Thomas D, Lopez JA, Martinez NR, Ellem KA, Schmidt CW (2007) Dendritic cell immunotherapy for stage IV melanoma. Melanoma Res 17:316–322
Ock CY, Hwang JE, Keam B, Kim SB, Shim JJ, Jang HJ, Park S, Sohn BH, Cha M, Ajani JA, Kopetz S, Lee KW, Kim TM, Heo DS, Lee JS (2017) Genomic landscape associated with potential response to anti-CTLA-4 treatment in cancers. Nat Commun 8:1050
Ott PA, Hu Z, Keskin DB, Shukla SA, Sun J, Bozym DJ, Zhang W, Luoma A, Giobbie-Hurder A, Peter L, Chen C, Olive O, Carter TA, Li S, Lieb DJ, Eisenhaure T, Gjini E, Stevens J, Lane WJ, Javeri I, Nellaiappan K, Salazar AM, Daley H, Seaman M, Buchbinder EI, Yoon CH, Harden M, Lennon N, Gabriel S, Rodig SJ, Barouch DH, Aster JC, Getz G, Wucherpfennig K, Neuberg D, Ritz J, Lander ES, Fritsch EF, Hacohen N, Wu CJ (2017) An immunogenic personal neoantigen vaccine for patients with melanoma. Nature 547:217–221
Pasetto A, Gros A, Robbins PF, Deniger DC, Prickett TD, Matus-Nicodemos R, Douek DC, Howie B, Robins H, Parkhurst MR, Gartner J, Trebska-McGowan K, Crystal JS, Rosenberg SA (2016) Tumor- and neoantigen-reactive T-cell receptors can be identified based on their frequency in fresh tumor. Cancer Immunol Res 4:734–743
Peters B, Sette A (2005) Generating quantitative models describing the sequence specificity of biological processes with the stabilized matrix method. BMC Bioinform 6:132
Peters B, Bulik S, Tampe R, Van Endert PM, Holzhutter HG (2003) Identifying MHC class I epitopes by predicting the TAP transport efficiency of epitope precursors. J Immunol 171:1741–1749
Pieper R, Christian RE, Gonzales MI, Nishimura MI, Gupta G, Settlage RE, Shabanowitz J, Rosenberg SA, Hunt DF, Topalian SL (1999) Biochemical identification of a mutated human melanoma antigen recognized by CD4(+) T cells. J Exp Med 189:757–766
Prickett TD, Crystal JS, Cohen CJ, Pasetto A, Parkhurst MR, Gartner JJ, Yao X, Wang R, Gros A, Li YF, El-Gamil M, Trebska-McGowan K, Rosenberg SA, Robbins PF (2016) Durable complete response from metastatic melanoma after transfer of autologous T cells recognizing 10 mutated tumor antigens. Cancer Immunol Res 4:669–678
Pritchard AL (2018) Targeting neoantigens for personalised immunotherapy. BioDrugs 32:99–109
Pritchard AL, Burel JG, Neller MA, Hayward NK, Lopez JA, Fatho M, Lennerz V, Wolfel T, Schmidt CW (2015a) Exome sequencing to predict neoantigens in melanoma. Cancer Immunol Res 3:992–998
Pritchard AL, Hastie ML, Neller M, Gorman JJ, Schmidt CW, Hayward NK (2015b) Exploration of peptides bound to MHC class I molecules in melanoma. Pigm Cell Melanoma Res 28:281–294
Purcell AW (2004) Isolation and characterization of naturally processed MHC-bound peptides from the surface of antigen-presenting cells. Methods Mol Biol 251:291–306
Purcell AW, Gorman JJ (2004) Immunoproteomics: mass spectrometry-based methods to study the targets of the immune response. Mol Cell Proteomics 3:193–208
Qian F, Gnjatic S, Jager E, Santiago D, Jungbluth A, Grande C, Schneider S, Keitz B, Driscoll D, Ritter G, Lele S, Sood A, Old LJ, Odunsi K (2004) Th1/Th2 CD4 + T cell responses against NY-ESO-1 in HLA-DPB1*0401/0402 patients with epithelial ovarian cancer. Cancer Immun 4:12
Rajasagi M, Shukla SA, Fritsch EF, Keskin DB, DeLuca D, Carmona E, Zhang W, Sougnez C, Cibulskis K, Sidney J, Stevenson K, Ritz J, Neuberg D, Brusic V, Gabriel S, Lander ES, Getz G, Hacohen N, Wu CJ (2014) Systematic identification of personal tumor-specific neoantigens in chronic lymphocytic leukemia. Blood 124:453–462
Reche PA, Glutting JP, Reinherz EL (2002) Prediction of MHC class I binding peptides using profile motifs. Hum Immunol 63:701–709
Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, Miller ML, Rekhtman N, Moreira AL, Ibrahim F, Bruggeman C, Gasmi B, Zappasodi R, Maeda Y, Sander C, Garon EB, Merghoub T, Wolchok JD, Schumacher TN, Chan TA (2015) Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science 348:124–128
Robbins PF, Morgan RA, Feldman SA, Yang JC, Sherry RM, Dudley ME, Wunderlich JR, Nahvi AV, Helman LJ, Mackall CL, Kammula US, Hughes MS, Restifo NP, Raffeld M, Lee CC, Levy CL, Li YF, El-Gamil M, Schwarz SL, Laurencot C, Rosenberg SA (2011) Tumor regression in patients with metastatic synovial cell sarcoma and melanoma using genetically engineered lymphocytes reactive with NY-ESO-1. J Clin Oncol 29:917–924
Robbins PF, Lu YC, El-Gamil M, Li YF, Gross C, Gartner J, Lin JC, Teer JK, Cliften P, Tycksen E, Samuels Y, Rosenberg SA (2013) Mining exomic sequencing data to identify mutated antigens recognized by adoptively transferred tumor-reactive T cells. Nat Med 19:747–752
Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N (2015) Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160:48–61
Rosati E, Dowds CM, Liaskou E, Henriksen EKK, Karlsen TH, Franke A (2017) Overview of methodologies for T-cell receptor repertoire analysis. BMC Biotechnol 17:61
Roskrow MA, Suzuki N, Gan Y, Sixbey JW, Ng CY, Kimbrough S, Hudson M, Brenner MK, Heslop HE, Rooney CM (1998) Epstein-Barr virus (EBV)-specific cytotoxic T lymphocytes for the treatment of patients with EBV-positive relapsed Hodgkin’s disease. Blood 91:2925–2934
Sahin U, Tureci O, Chen YT, Seitz G, Villena-Heinsen C, Old LJ, Pfreundschuh M (1998) Expression of multiple cancer/testis (CT) antigens in breast cancer and melanoma: basis for polyvalent CT vaccine strategies. Int J Cancer 78:387–389
Sahin U, Derhovanessian E, Miller M, Kloke BP, Simon P, Lower M, Bukur V, Tadmor AD, Luxemburger U, Schrors B, Omokoko T, Vormehr M, Albrecht C, Paruzynski A, Kuhn AN, Buck J, Heesch S, Schreeb KH, Muller F, Ortseifer I, Vogler I, Godehardt E, Attig S, Rae R, Breitkreuz A, Tolliver C, Suchan M, Martic G, Hohberger A, Sorn P, Diekmann J, Ciesla J, Waksmann O, Bruck AK, Witt M, Zillgen M, Rothermel A, Kasemann B, Langer D, Bolte S, Diken M, Kreiter S, Nemecek R, Gebhardt C, Grabbe S, Holler C, Utikal J, Huber C, Loquai C, Tureci O (2017) Personalized RNA mutanome vaccines mobilize poly-specific therapeutic immunity against cancer. Nature 547:222–226
Schmidt J, Guillaume P, Dojcinovic D, Karbach J, Coukos G, Luescher I (2017) In silico and cell-based analyses reveal strong divergence between prediction and observation of T-cell-recognized tumor antigen T-cell epitopes. J Biol Chem 292:11840–11849
Schrors B, Lubcke S, Lennerz V, Fatho M, Bicker A, Wolfel C, Derigs P, Hankeln T, Schadendorf D, Paschen A, Wolfel T (2017) HLA class I loss in metachronous metastases prevents continuous T cell recognition of mutated neoantigens in a human melanoma model. Oncotarget 8:28312–28327
Schuessler A, Smith C, Beagley L, Boyle GM, Rehan S, Matthews K, Jones L, Crough T, Dasari V, Klein K, Smalley A, Alexander H, Walker DG, Khanna R (2014) Autologous T-cell therapy for cytomegalovirus as a consolidative treatment for recurrent glioblastoma. Cancer Res 74:3466–3476
Schuler MM, Nastke MD, Stevanovikc S (2007) SYFPEITHI: database for searching and T-cell epitope prediction. Methods Mol Biol 409:75–93
Schumacher TN, Schreiber RD (2015) Neoantigens in cancer immunotherapy. Science 348:69–74
Segal NH, Parsons DW, Peggs KS, Velculescu V, Kinzler KW, Vogelstein B, Allison JP (2008) Epitope landscape in breast and colorectal cancer. Cancer Res 68:889–892
Sharkey MS, Lizee G, Gonzales MI, Patel S, Topalian SL (2004) CD4(+) T-cell recognition of mutated B-RAF in melanoma patients harboring the V599E mutation. Cancer Res 64:1595–1599
Sharma P, Kranz DM (2016) Recent advances in T-cell engineering for use in immunotherapy. F1000Research 5
Shono Y, Tanimura H, Iwahashi M, Tsunoda T, Tani M, Tanaka H, Matsuda K, Yamaue H (2003) Specific T-cell immunity against Ki-ras peptides in patients with pancreatic and colorectal cancers. Br J Cancer 88:530–536
Siebenkas C, Chiappinelli KB, Guzzetta AA, Sharma A, Jeschke J, Vatapalli R, Baylin SB, Ahuja N (2017) Inhibiting DNA methylation activates cancer testis antigens and expression of the antigen processing and presentation machinery in colon and ovarian cancer cells. PLoS ONE 12:e0179501
Singh SP, Mishra BN (2008) Prediction of MHC binding peptide using Gibbs motif sampler, weight matrix and artificial neural network. Bioinformation 3:150–155
Skipper JC, Hendrickson RC, Gulden PH, Brichard V, Van Pel A, Chen Y, Shabanowitz J, Wolfel T, Slingluff CL Jr, Boon T, Hunt DF, Engelhard VH (1996) An HLA-A2-restricted tyrosinase antigen on melanoma cells results from posttranslational modification and suggests a novel pathway for processing of membrane proteins. J Exp Med 183:527–534
Smith C, Lee V, Schuessler A, Beagley L, Rehan S, Tsang J, Li V, Tiu R, Smith D, Matthews MAN, Gostick KK, Price E, Burrows DA, Boyle J, Chua GM, Panizza D, Porceddu B, Nicholls SV, Kwong DLW, Khanna R (2017) Pre-emptive and therapeutic adoptive immunotherapy for nasopharyngeal carcinoma: Phenotype and effector function of T cells impact on clinical response. Oncoimmunology 6:e1273311
Smith-Garvin JE, Koretzky GA, Jordan MS (2009) T cell activation. Annu Rev Immunol 27:591–619
Snyder A, Makarov V, Merghoub T, Yuan J, Zaretsky JM, Desrichard A, Walsh LA, Postow MA, Wong P, Ho TS, Hollmann TJ, Bruggeman C, Kannan K, Li Y, Elipenahli C, Liu C, Harbison CT, Wang L, Ribas A, Wolchok JD, Chan TA (2014) Genetic basis for clinical response to CTLA-4 blockade in melanoma. N Engl J Med 371:2189–2199
Somasundaram R, Swoboda R, Caputo L, Otvos L, Weber B, Volpe P, van Belle P, Hotz S, Elder DE, Marincola FM, Schuchter L, Guerry D, Czerniecki BJ, Herlyn D (2006) Human leukocyte antigen-A2-restricted CTL responses to mutated BRAF peptides in melanoma patients. Cancer Res 66:3287–3293
Stern LJ, Brown JH, Jardetzky TS, Gorga JC, Urban RG, Strominger JL, Wiley DC (1994) Crystal structure of the human class II MHC protein HLA-DR1 complexed with an influenza virus peptide. Nature 368:215–221
Strebhardt K, Ullrich A (2008) Paul Ehrlich’s magic bullet concept: 100 years of progress. Nature Rev Cancer 8:473–480
Stronen E, Toebes M, Kelderman S, van Buuren MM, Yang W, van Rooij N, Donia M, Boschen ML, Lund-Johansen F, Olweus J, Schumacher TN (2016) Targeting of cancer neoantigens with donor-derived T cell receptor repertoires. Science 352:1337–1341
Sun Z, Chen F, Meng F, Wei J, Liu B (2017) MHC class II restricted neoantigen: a promising target in tumor immunotherapy. Cancer Lett 392:17–25
Tan CT, Croft NP, Dudek NL, Williamson NA, Purcell AW (2011) Direct quantitation of MHC-bound peptide epitopes by selected reaction monitoring. Proteomics 11:2336–2340
Tan MP, Gerry AB, Brewer JE, Melchiori L, Bridgeman JS, Bennett AD, Pumphrey NJ, Jakobsen BK, Price DA, Ladell K, Sewell AK (2015) T cell receptor binding affinity governs the functional profile of cancer-specific CD8 + T cells. Clin Exp Immunol 180:255–270
Tappeiner E, Finotello F, Charoentong P, Mayer C, Rieder D, Trajanoski Z (2017) TIminer: NGS data mining pipeline for cancer immunology and immunotherapy. Bioinformatics 33:3140–3141
Tashiro H, Brenner MK (2017) Immunotherapy against cancer-related viruses. Cell Res 27:59–73
Tenzer S, Peters B, Bulik S, Schoor O, Lemmel C, Schatz MM, Kloetzel PM, Rammensee HG, Schild H, Holzhutter HG (2005) Modeling the MHC class I pathway by combining predictions of proteasomal cleavage, TAP transport and MHC class I binding. Cell Mol Life Sci 62:1025–1037
Toes RE, Nussbaum AK, Degermann S, Schirle M, Emmerich NP, Kraft M, Laplace C, Zwinderman A, Dick TP, Muller J, Schonfisch B, Schmid C, Fehling HJ, Stevanovic S, Rammensee HG, Schild H (2001) Discrete cleavage motifs of constitutive and immunoproteasomes revealed by quantitative analysis of cleavage products. J Exp Med 194:1–12
Tran E, Turcotte S, Gros A, Robbins PF, Lu YC, Dudley ME, Wunderlich JR, Somerville RP, Hogan K, Hinrichs CS, Parkhurst MR, Yang JC, Rosenberg SA (2014) Cancer immunotherapy based on mutation-specific CD4 + T cells in a patient with epithelial cancer. Science 344:641–645
Tran E, Ahmadzadeh M, Lu YC, Gros A, Turcotte S, Robbins PF, Gartner JJ, Zheng Z, Li YF, Ray S, Wunderlich JR, Somerville RP, Rosenberg SA (2015) Immunogenicity of somatic mutations in human gastrointestinal cancers. Science 350:1387–1390
Tran E, Robbins PF, Lu YC, Prickett TD, Gartner JJ, Jia L, Pasetto A, Zheng Z, Ray S, Groh EM, Kriley IR, Rosenberg SA (2016) T-cell transfer therapy targeting mutant KRAS in cancer. N Engl J Med 375:2255–2262
Tsuji T, Altorki NK, Ritter G, Old LJ, Gnjatic S (2009) Characterization of preexisting MAGE-A3-specific CD4 + T cells in cancer patients and healthy individuals and their activation by protein vaccination. J Immunol 183:4800–4808
Tynan FE, Burrows SR, Buckle AM, Clements CS, Borg NA, Miles JJ, Beddoe T, Whisstock JC, Wilce MC, Silins SL, Burrows JM, Kjer-Nielsen L, Kostenko L, Purcell AW, McCluskey J, Rossjohn J (2005) T cell receptor recognition of a ‘super-bulged’ major histocompatibility complex class I-bound peptide. Nat Immunol 6:1114–1122
Uebel S, Tampe R (1999) Specificity of the proteasome and the TAP transporter. Curr Opin Immunol 11:203–208
van Rooij N, van Buuren MM, Philips D, Velds A, Toebes M, Heemskerk B, van Dijk LJ, Behjati S, Hilkmann H, El Atmioui D, Nieuwland M, Stratton MR, Kerkhoven RM, Kesmir C, Haanen JB, Kvistborg P, Schumacher TN (2013) Tumor exome analysis reveals neoantigen-specific T-cell reactivity in an ipilimumab-responsive melanoma. J Clin Oncol 31:e439–e442
Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA (2015) Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350:207–211
van der Bruggen P, Traversari C, Chomez P, Lurquin C, De Plaen E, Van den Eynde B, Knuth A, Boon T (1991) A gene encoding an antigen recognized by cytolytic T lymphocytes on a human melanoma. Science 254:1643–1647
Veatch JR, Lee SM, Fitzgibbon M, Chow IT, Jesernig B, Schmitt T, Kong YY, Kargl J, Houghton AM, Thompson JA, McIntosh M, Kwok WW, Riddell SR (2018) Tumor-infiltrating BRAFV600E-specific CD4 + T cells correlated with complete clinical response in melanoma. J Clin Invest 128:1563–1568
Vella LA, Yu M, Fuhrmann SR, El-Amine M, Epperson DE, Finn OJ (2009) Healthy individuals have T-cell and antibody responses to the tumor antigen cyclin B1 that when elicited in mice protect from cancer. Proc Natl Acad Sci USA 106:14010–14015
Verdegaal EM, de Miranda NF, Visser M, Harryvan T, van Buuren MM, Andersen RS, Hadrup SR, van der Minne CE, Schotte R, Spits H, Haanen JB, Kapiteijn EH, Schumacher TN, van der Burg SH (2016) Neoantigen landscape dynamics during human melanoma-T cell interactions. Nature 536:91–95
Vigneron N, Stroobant V, Van den Eynde BJ, van der Bruggen P (2013) Database of T cell-defined human tumor antigens: the 2013 update. Cancer Immunity 13:15
Vita R, Zarebski L, Greenbaum JA, Emami H, Hoof I, Salimi N, Damle R, Sette A, Peters B (2010) The immune epitope database 2.0. Nucleic Acids Res 38:D854–D862
Walboomers JM, Jacobs MV, Manos MM, Bosch FX, Kummer JA, Shah KV, Snijders PJ, Peto J, Meijer CJ, Munoz N (1999) Human papillomavirus is a necessary cause of invasive cervical cancer worldwide. J Pathol 189:12–19
Wang RF, Wang X, Atwood AC, Topalian SL, Rosenberg SA (1999) Cloning genes encoding MHC class II-restricted antigens: mutated CDC27 as a tumor antigen. Science 284:1351–1354
Wang Y, Wu XJ, Zhao AL, Yuan YH, Chen YT, Jungbluth AA, Gnjatic S, Santiago D, Ritter G, Chen WF, Old LJ, Ji JF (2004) Cancer/testis antigen expression and autologous humoral immunity to NY-ESO-1 in gastric cancer. Cancer Immun 4:11
Wang P, Sidney J, Dow C, Mothe B, Sette A, Peters B (2008) A systematic assessment of MHC class II peptide binding predictions and evaluation of a consensus approach. PLoS Comput Biol 4:e1000048
Wang K, Wei G, Liu D (2012) CD19: a biomarker for B cell development, lymphoma diagnosis and therapy. Exp Hematol Oncol 1:36
Wang Y, Lin Y, Shu M, Wang R, Hu Y, Lin Z (2013) Proteasomal cleavage site prediction of protein antigen using BP neural network based on a new set of amino acid descriptor. J Mol Model 19:3045–3052
White MK, Pagano JS, Khalili K (2014) Viruses and human cancers: a long road of discovery of molecular paradigms. Clin Microbiol Rev 27:463–481
Wucherpfennig KW, Gagnon E, Call MJ, Huseby ES, Call ME (2010) Structural biology of the T-cell receptor: insights into receptor assembly, ligand recognition, and initiation of signaling. Cold Spring Harb Perspect Biol 2:a005140
Yadav M, Jhunjhunwala S, Phung QT, Lupardus P, Tanguay J, Bumbaca S, Franci C, Cheung TK, Fritsche J, Weinschenk T, Modrusan Z, Mellman I, Lill JR, Delamarre L (2014) Predicting immunogenic tumour mutations by combining mass spectrometry and exome sequencing. Nature 515:572–576
Yu Z, Theoret MR, Touloukian CE, Surman DR, Garman SC, Feigenbaum L, Baxter TK, Baker BM, Restifo NP (2004) Poor immunogenicity of a self/tumor antigen derives from peptide-MHC-I instability and is independent of tolerance. J Clin Investig 114:551–559
Yuen GJ, Demissie E, Pillai S (2016) B lymphocytes and cancer: a love-hate relationship. Trends Cancer 2:747–757
Zacharakis N, Chinnasamy H, Black M, Xu H, Lu YC, Zheng Z, Pasetto A, Langhan M, Shelton T, Prickett T, Gartner J, Jia L, Trebska-McGowan K, Somerville RP, Robbins PF, Rosenberg SA, Goff SL, Feldman SA (2018) Immune recognition of somatic mutations leading to complete durable regression in metastatic breast cancer. Nat Med 24:724–730
Zendman AJ, de Wit NJ, van Kraats AA, Weidle UH, Ruiter DJ, van Muijen GN (2001) Expression profile of genes coding for melanoma differentiation antigens and cancer/testis antigens in metastatic lesions of human cutaneous melanoma. Melanoma Res 11:451–459
Zendman AJ, Ruiter DJ, Van Muijen GN (2003) Cancer/testis-associated genes: identification, expression profile, and putative function. J Cell Physiol 194:272–288
Zhang GL, Petrovsky N, Kwoh CK, August JT, Brusic V (2006) PRED(TAP): a system for prediction of peptide binding to the human transporter associated with antigen processing. Immunome Res 2:3
Zhang H, Lund O, Nielsen M (2009) The PickPocket method for predicting binding specificities for receptors based on receptor pocket similarities: application to MHC-peptide binding. Bioinformatics 25:1293–1299
Zhu Y, Rudensky AY, Corper AL, Teyton L, Wilson IA (2003) Crystal structure of MHC class II I-Ab in complex with a human CLIP peptide: prediction of an I-Ab peptide-binding motif. J Mol Biol 326:1157–1174
Acknowledgements
This review brings together a large amount of work in a rapidly developing research area, in order to provide an overall picture of the current state of the field. As a result, there are recommendations throughout to further primary research and review papers on a specific topic. We apologise to our colleagues who may feel their work is inadequately cited, or that further details should have been provided here. SH and ALP are supported by Highland and Island Enterprise, Scotland (HMS9353763).
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Hutchison, S., Pritchard, A.L. Identifying neoantigens for use in immunotherapy. Mamm Genome 29, 714–730 (2018). https://doi.org/10.1007/s00335-018-9771-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-018-9771-6