Abstract
Immunotherapy has changed the landscape of cancer treatment. The introduction of immune checkpoint inhibitors has seen tremendous success in improving overall survival of patients with advanced metastatic cancers and has now become the standard of care for multiple tumor types. However, efficacy of immune checkpoint blockade appears to be limited to immunogenic cancers, and even amongst immune-reactive cancers, response rates are low and variable between patients. Recent data have also demonstrated the rapid emergence of resistance to immune checkpoint inhibitors, with some patients progressing on treatment within one year. Significant research efforts are now directed at identifying predictive biomarkers and mechanisms of resistance to immune checkpoint blockade. These studies are underpinned by comprehensive and detailed profiling of the immune milieu. In this review, we discuss the utility and efficacy of immune cell profiling to uncover biomarkers of response and mechanisms of resistance to immune checkpoint inhibitors.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
One of the hallmarks of cancer is the evasion of immune surveillance, arising from the improper monitoring of malignant cells by the immune system due to alterations in oncogenic signaling pathways or changes in the local microenvironment. Cancer cells can downregulate expression of antigens and antigen presentation molecules to hinder immune cell recognition, and conversely, promote expression of immunosuppressive molecules to dampen anti-tumor immune activity. Thus, cancer cells tip the balance towards immune evasion, enabling cancer development and progression (Chen and Mellman 2013, 2017; Vinay et al. 2015; Muenst et al. 2016).
Given that cancers propagate due to dysfunctional immune recognition and activity, several immune-based therapies or immunotherapies that boost immune responses against cancer have been developed. Cytokines such as interferon-alpha2b and interleukin-2 promote cytotoxic T and natural killer (NK) cell activity, and were approved for the treatment of high-risk metastatic melanoma in 1996 and 1998, respectively (Bhatia et al. 2009). The dendritic cell vaccine sipuleucel-T, approved for the treatment of stage IV metastatic prostate cancer, induces cytotoxic T cell responses and led to a 4-month improvement in median overall survival (Kantoff et al. 2010). Other types of vaccines using cancer antigens and adjuvant tumor lysates have been tested in clinical trials with varying efficacy in different cancer types [reviewed in (Finn 2003; Melief et al. 2015; van der Burg et al. 2016)]. Adoptive cell transfer (ACT) (Restifo et al. 2012; Yang and Rosenberg 2016), including chimeric antigen receptor (CAR) T cell therapy (Ramos et al. 2016; Newick et al. 2017), involving the extraction and manipulation of patients’ immune cells, has also improved response rates and survival in certain cancer types.
Amongst the different types of immunotherapies, immune checkpoint inhibitors targeting cytotoxic T lymphocyte-associated antigen 4 (CTLA-4) or programmed death-1/programmed death-ligand 1 (PD-1/PD-L1) signaling have received significant attention in the past 5 years. Under normal conditions, these inhibitory immune checkpoints suppress T cell activity to counteract overactivation of the immune response, and prevent excessive inflammation and tissue damage. However, elevated expression of these inhibitory checkpoints in cancer inhibits anti-tumor T cell function, and immune checkpoint inhibitors are able to mitigate these suppressive effects [reviewed in (Pardoll 2012; Topalian et al. 2015)]. Immune checkpoint inhibitors against CTLA-4, PD-1, and PD-L1 have now been approved by the US Food and Drug Administration (FDA) for the treatment of different cancer types (Table 1).
Immune checkpoint inhibitors have shown most success in the treatment of advanced metastatic melanoma (McDermott et al. 2014). Treatment with anti-CTLA-4 benefits approximately 20% of patients with advanced melanoma, and a small proportion of these patients remain disease free 10 years after treatment (Schadendorf et al. 2015). Objective response rates are higher with anti-PD-1 treatment (44%) (Robert et al. 2015), and although the combination of anti-CTLA-4 and anti-PD-1 enhances response rate to 58% (Larkin et al. 2015), combination therapy leads to significant treatment-related toxicities (Hodi et al. 2016). Resistance to immune checkpoint inhibitors is an emerging problem, with almost 50% of anti-PD-1-treated patients showing progression within 1 year (Daud et al. 2015).
The limitations of immune checkpoint inhibitors necessitate the identification of biomarkers that can help prognosticate the disease and predict treatment response to improve patient selection and management. Moreover, there is an urgent need to better understand the biological processes underlying the development of resistance to immune checkpoint blockade. In this review, we discuss how profiling of the immune cell contexture can contribute to the discovery of biomarkers and the mechanistic understanding of resistance to immune checkpoint inhibitors.
Immune cell profiling to uncover biomarkers of response
Cancer biomarkers can provide information on the presence and stage of disease, disease progression and outcomes, and the efficacy and benefits of particular therapies (Kulasingam and Diamandis 2008). Biomarkers can be detected and measured in tissues or blood, and given the importance of immune cell activity in immune checkpoint therapies, there is now a concerted effort to profile and characterize immune cells in tissue and blood biopsies to discover potential biomarkers of response.
Immune cell counts and proximity
Presence of immune cells can be assessed in tissue biopsies by immunohistochemical (IHC) staining with hematoxylin and eosin, or with antibodies against specific cell surface receptors that are conjugated to different chromogens or fluorescence (Matos et al. 2010). The development of multispectral imaging and detection systems has now enabled multiplex IHC that can detect multiple protein antigens, and differentiate distinct immune cell types in a single tumor sample (Stack et al. 2014; Dixon et al. 2015). This technique offers the added advantage of providing architectural and spatial information about immune cells within the tumor microenvironment. The immune contexture of tumors, encompassing the density, composition, functional state, and organization of immune cells in relation to the tumor, has been shown to provide vital information on disease progression, prognosis, and prediction of treatment response (Fridman et al. 2012, 2017). Indeed, the immunoscore, derived from the density of intratumoral and peritumoral T cells, was shown to be a better prognostic marker in colorectal cancer staging than the tumor, node, and metastasis (TNM) system (Galon et al. 2006, 2014). The immunoscore has also shown prognostic value in other cancer types including breast, prostate, kidney, lung, and melanoma [reviewed in (Galon et al. 2012, 2014; Becht et al. 2016a, b)] and efforts are underway to evaluate its utility in predicting treatment response in other cancer types (Ascierto et al. 2013; Becht et al. 2016a, b). Additionally, although enumeration of T lymphocytes has prognostic significance, identification of specific immune cell subsets is equally important. For example, higher frequencies of CD8 tumor infiltrating T cells are associated with improved clinical outcome and survival (Zhang et al. 2003; Sato et al. 2005), but the presence of T regulatory cells (CD4+ FoxP3+) in tumor tissues is associated with reduced survival and high death hazard in patients with ovarian carcinoma (Curiel et al. 2004).
In melanoma patients, increase in tumor infiltrating lymphocytes (TILs) 3 weeks after treatment with anti-CTLA-4 was associated with clinical activity. Interestingly, high baseline expression of FoxP3, a marker of T regulatory cells, was also associated with clinical activity but expression of CD4, CD8, granzyme B and perforin, markers of effector cytotoxic T cells, did not correlate with response (Hamid et al. 2011). Analysis of CD8 T cell density was also performed on tumor biopsies from 46 melanoma patients receiving anti-PD-1 therapy, at baseline and during treatment. In contrast, intra- and peri-tumoral CD8 T cell density was associated with anti-PD-1 response with AUC ROC (area under the receiver operating characteristic curve) value of > 0.9 (Tumeh et al. 2014). In a separate study, Vilain et al. confirmed that patients responding to anti-PD-1 treatment displayed higher baseline intra-tumoral and peri-tumoral CD8 T cell counts and differences in CD8 T cell counts between responding and non-responding patients were more pronounced in early on treatment biopsies (Vilain et al. 2017). Similarly, Chen et al. also showed significantly higher CD8 T cell density in baseline and particularly, in early on treatment biopsies from melanoma patients responding to sequential anti-CTLA-4 and anti-PD-1 therapy (Chen et al. 2016). Collectively, these studies demonstrate that enumeration and spatiotemporal analysis of CD8 T cells may represent useful biomarkers of response to immune checkpoint blockade.
Characterisation of immune cell subsets and phenotypes
Although IHC provides structural and spatial information, analysis is restricted to tissue biopsies and this technique alone cannot fully characterize the different immune cell types present in tissues. This is especially limiting for analysis of blood biopsies considering the many different subsets of immune cells circulating in blood. A differential blood count can be used to assess numbers and proportion of white blood cells (typically neutrophils, lymphocytes, monocytes, eosinophils, and basophils), and have been applied to diagnose hematologic malignancies and immune system disorders (Blumenreich 1990). Several recent studies have demonstrated the utility of differential blood count analysis to identify predictive biomarkers of treatment response. For example, low absolute monocyte counts and high absolute eosinophil counts at baseline were associated with a favorable outcome in melanoma patients (n = 209) treated with anti-CTLA-4 (Martens et al. 2016). Similarly, high baseline counts of eosinophils and lymphocytes were associated with increased overall survival in melanoma patients (n = 616) treated with anti-PD-1 (Weide et al. 2016). Baseline absolute leukocyte and neutrophil counts were also shown to be prognostic in 65 patients with non-small cell lung carcinoma (NSCLC) treated with anti-PD-1 or anti-PD-L1. Specifically, the neutrophil to lymphocyte ratio significantly correlated with survival, and increase in this ratio was prognostic for shorter survival (Mezquita et al. 2017).
There are now mature multiparameter detection systems including flow and mass cytometry that enable the concurrent phenotypic and functional characterization of many different immune cell subsets (Perfetto et al. 2004; Ornatsky et al. 2010). These technologies can be used to investigate immune cell profiles in blood and tissue biopsies, the latter requiring prior processing and disaggregation of tissues into single cells. Flow cytometry uses antibodies that are conjugated to different fluorochromes, and the latest flow cytometer systems allow interrogation of up to 23 parameters using six lasers and 21 florescent detectors (Perfetto et al. 2004). However, flow cytometry is limited by spectral overlap of the fluorescent dyes, whereas mass cytometry overcomes this limitation using antibodies conjugated to heavy metal ions that are subsequently detected by mass spectrometry. Mass cytometry has surpassed the multiplexing capacity of flow cytometry, measuring up to 40 different parameters (Bendall et al. 2012; Spitzer and Nolan 2016), thus allowing the discrimination of different immune cell subsets and phenotypic characterization of their activation, proliferation, and tolerance states. For example, expression of immune exhaustion markers (e.g., PD-1, LAG3, TIM3) may indicate T cell anergy (Baitsch et al. 2011), and analysis of transcription factors and phosphorylated proteins following cell permeabilisation can provide additional information on signaling pathways in immune cells. These cytometry platforms can also incorporate identification of peptide-MHC multimers to characterize T cell reactivity, and cell cycle analysis to determine cell proliferation and viability [reviewed in (Krutzik et al. 2004; Covey and Cesano 2010; Andersen et al. 2012; Bjornson et al. 2013; Newell 2013)]. Advanced analytical platforms such as viSNE (Amir el et al. 2013) and SPADE (Qiu et al. 2011) are now available to allow phenotypic visualization of the high-dimensional single-cell data generated from flow and mass cytometry.
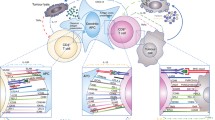
Standardized panels for whole blood immune cell phenotyping using flow or mass cytometry have been proposed and used for the diagnosis of blood-based cancers such as leukemia (Maecker et al. 2012; van Dongen et al. 2012). These panels can be similarly applied and modified to evaluate major populations of distinct immune cell subsets in blood and tissues of patients treated with immune checkpoint inhibitors to uncover potential cell-based biomarkers (Fig. 1). For instance, flow cytometric analysis of peripheral blood mononuclear cells (PBMCs) from 36 metastatic melanoma patients revealed increased frequency of ICOS + CD4 T cells following treatment with anti-CTLA-4, suggesting that this effector T cell population may function as a pharmacodynamic marker of anti-CTLA-4 therapy (Ng Tang et al. 2013). Comparison of PBMC profiles from melanoma patients treated with anti-PD-1 or anti-CTLA-4 showed distinct frequencies of immune cell subsets that may serve as predictive biomarkers. For example, CD4 and CD8 memory T cells were enriched in patients who responded to anti-CTLA-4 therapy whereas increased CD69 + NK cell counts correlated with clinical response to anti-PD-1 therapy (Subrahmanyam et al. 2018). Further, flow cytometric profiling of PBMCs from 29 stage IV melanoma patients before and after treatment with anti-PD-1 identified a reinvigorated exhausted Ki67 + CD8 T cell population. A ratio greater than 1.94 of this CD8 T cell subset to tumor burden significantly correlated with better objective response, progression free survival, and overall survival (Huang et al. 2017). Similarly, mass cytometric analysis of PBMCs from 20 melanoma patients at baseline and 12 weeks after anti-PD-1 therapy revealed frequency of a monocytic population (CD14+ CD16− HLA-DRhi) to be a strong predictor of progression free and overall survival (Krieg et al. 2018).
Proposed flow and mass cytometry marker panels for the identification and characterization of major immune cell subsets, including monocytes, granulocytes, natural killer, dendritic, B and T cells. G-MDSC denotes granulocytic myeloid-derived suppressor cell while Mo-MDSC denotes monocytic myeloid-derived suppressor cell
Flow or mass cytometric profiling can also be performed on tissue biopsies to expose potential biomarkers. Daud et al. showed significant correlation between CD8 T cells expressing high levels of CTLA-4 and PD-1 with clinical response and progression free survival in freshly isolated metastatic melanoma samples from 40 patients prior to treatment with anti-PD-1, suggesting that relative abundance of this T cell population may predict treatment response (Daud et al. 2016). It is worth mentioning that there may be differences in immune cell counts in peripheral blood compared to the tissue microenvironment, and these differences may reflect immune cell egress from circulation into the tumor tissue.
Transcriptomic biomarkers of immune cells
Transcriptomic profiling of immune cells in fresh frozen and paraffin embedded tissues using RNA sequencing, microarrays, or the NanoString nCounter platform has provided comprehensive gene expression data on immune cell types and abundance, molecular mechanisms of immune cell activation and behavior, and the inflammatory state of the tumor microenvironment.
Expressions of a specific gene or gene sets similar to the gene panels offered by NanoString (Table 2) are used to differentiate immune cell subsets, including lymphocytic T and B cells, natural killer and dendritic cells, and myeloid cells comprising of neutrophils, macrophages, and eosinophils. However, transcriptome data are derived from heterogeneous cell types in the tumor including malignant, stromal, and immune cells. Several computational algorithms have been designed to deconvolute gene expression data from complex tissue biopsies to estimate immune cell frequencies [reviewed in (Finotello and Trajanoski 2018)]. Programs such as TIMER (Li et al. 2016) and CIBERSORT (Newman et al. 2015) are publicly available and provide estimations of the composition and degree of immune cell infiltration.
In addition to immune cell abundance, immune phenotypes can also be discerned based on expression of specific cytokines and mediators (Becker et al. 2015), and the Immunological Genome Project recently aimed to decipher the signaling networks of 249 immune cell types to uncover molecular pathways involved in immune response and interaction (Heng et al. 2008). Combination of genes has also been used to infer immune activity. For example, the geometric mean expression of PRF1 and GZMA transcripts was shown to correlate with cytolytic activity of immune infiltrates, and this cytolytic (CYT) score associated with survival benefit in a range of cancer types (Rooney et al. 2015). Several immune cell signatures that reflect immune differentiation, activation, and signaling have also been proposed (Shaffer et al. 2001; Critchley-Thorne et al. 2011; Godec et al. 2016). Expression of these immune response gene sets, which include antigen presentation molecules (i.e., major histocompatibility complex molecules), interferon signaling effectors, T cell activation, adaptive and innate immunity genes was shown to correlate with prolonged survival in metastatic melanoma patients (Mandruzzato et al. 2006; Bogunovic et al. 2009), relapse free survival in patients with small cell lung cancer (Roepman et al. 2009), and extended time to relapse and recurrence in colon cancer patients (Galon et al. 2006).
In patients treated with immune checkpoint inhibitors, gene expression profiles and signatures reflective of an active immune microenvironment have been shown to correlate with clinical activity [reviewed in (Gajewski et al. 2010; Ulloa-Montoya et al. 2013)], and may serve as biomarkers of treatment response. For example, transcriptome analysis of tumor biopsies from 40 melanoma patients before treatment with anti-CTLA-4 indicated higher expression of the CYT score, CTLA-4, PD-1, PD-L1, and PD-L2 in patients with clinical benefit (Van Allen et al. 2015). Similarly, baseline expression of immune-associated genes including T cell surface markers (CD8, CD3, CD38), cytokines involved in T cell recruitment (CXCL9 and CXCL10), immune receptors (CXCR6 and CCR5), and TNF signaling components correlated with response to anti-CTLA-4 therapy and overall survival, and these associations were more pronounced in early on treatment biopsies (3 weeks after treatment initiation) (Ji et al. 2012). Transcriptomic profiling of longitudinal tumor biopsies allows investigation into the dynamics of immune response during treatment, and in a cohort of melanoma patients treated with anti-PD-1 (n = 45), comparison of baseline to on treatment biopsies revealed increased induction of immune-related genes in patients with clinical benefit (Riaz et al. 2017). Similarly, Chen et al. showed significant upregulation of genes associated with IFNγ signaling, antigen presentation, and T cell effector function in on treatment biopsies from 54 melanoma patients treated with sequential anti-CTLA-4 and anti-PD-1 therapies (Chen et al. 2016).
The IFNγ signaling pathway regulates numerous aspects of the immune response, including expression of immune checkpoint proteins, antigen processing and presentation molecules, and various chemokines involved in immune cell recruitment. Several components of the IFNγ signaling pathway including STAT1, STAT2, STAT3, and IRF1 were upregulated in anti-PD-1 responding tumors (Garcia-Diaz et al. 2017), and loss of function mutations in JAK1/JAK2 have been implicated to confer resistance to anti-PD-1 therapy (Zaretsky et al. 2016; Shin et al. 2017). As such, expression of IFNγ pathway signatures may represent surrogate markers of immune activity and response to immune checkpoint therapy. Indeed, high IFNγ expression was shown to be associated with longer progression free survival in non-small cell lung cancer (NSCLC) and melanoma patients treated with anti-PD-1 (Karachaliou et al. 2018). In keeping with this, expression of a 10-gene IFNγ signature was initially interrogated in baseline tumor biopsies from 19 melanoma patients treated with anti-PD-1. Expression of a 28-gene expanded immune signature that incorporated the 10 IFNγ genes as well as T cell and antigen presentation molecules was further validated in a separate cohort of 62 melanoma patients. Both these signatures showed significant association with overall response rate and progression free survival (Ribas et al. 2016), and when examined in baseline tumor samples from head and neck squamous cell carcinoma (HNSCC; n = 43) and gastric cancer (n = 33) patients treated with anti-PD-1, these signatures also correlated with overall response rate and progression free survival (Ayers et al. 2017). A similar T effector IFNγ gene signature was investigated in NSCLC patients treated with anti-PD-L1 (n = 142), and high expression of the signature at baseline correlated with improved overall survival (Fehrenbacher et al. 2016). However, it is important to mention that expression of IFNγ-associated signatures do not always associate with clinical benefit. For example, the T effector IFNγ signature showed no association with clinical benefit in 70 renal cell carcinoma patients treated with anti-PD-L1 (McDermott et al. 2016).
Recently, an IPRES signature, comprised of genes involved in mesenchymal transition, extracellular matrix remodeling, angiogenesis, and wound healing, was shown to be enriched in baseline biopsies of melanoma patients with innate resistance to anti-PD-1 therapy (Hugo et al. 2017). Although co-enrichment of this signature was identified across other cancer types (Hugo et al. 2017), its expression was not associated with innate resistance to anti-CTLA-4 therapy, and thus far, correlation of the IPRES signature with resistance has not been found in other cohorts of patients treated with immune checkpoint blockade (Van Allen et al. 2015; Chen et al. 2016; Riaz et al. 2017).
Transcription profiling can also be performed on homogenous cells instead of admix tissue biopsies. Immune cells can be first isolated from tumor tissues using laser capture microdissection, or tumor dissociation followed by single cell sorting and immune cell enrichment before RNA sequencing. A recent study by Tirosh et al. conducted single-cell RNA sequencing on tumor, immune, and stromal cells isolated from melanoma tumors and reported distinct T cell functional states from the isolated immune cells (Tirosh et al. 2016). Sequencing of the T cell receptor can also be performed to identify T cell repertoire and clonal expansion (Postow et al. 2015; Riaz et al. 2017). Riaz et al. identified temporal changes in the intratumoral TCR repertoire indicative of T cell clonal expansion during anti-PD-1 treatment. In melanoma patients who have progressed on anti-CTLA-4 therapy and subsequently treated with anti-PD-1 (n = 35), an increase in the number of unique CDR3 sequences (richness) was associated with clinical benefit (Riaz et al. 2017). Additionally, Postow et al. showed that increased TCR repertoire and T cell diversity in peripheral blood of 12 melanoma patients before anti-CTLA-4 therapy was associated with clinical benefit (Postow et al. 2015).
Immune cell profiling to elucidate resistance mechanisms
Resistance to immune checkpoint inhibitors may be innate, whereby patients do not respond to therapy at first instance, or acquired, in which patients responded initially but subsequently progress. Innate and acquired resistance mechanisms can be tumor intrinsic, involving changes in the cancer cells, or tumor extrinsic, attributed to alterations in the tumor micro-environment (Sharma et al. 2017). Tumor intrinsic mechanisms include downregulation or loss of antigens and antigen presentation molecules (Zaretsky et al. 2016), disruption in the IFNγ signaling pathway (Zaretsky et al. 2016; Garcia-Diaz 2017) and T cell exclusion (Liu et al. 2013; Peng et al. 2016). Tumor extrinsic mechanisms involve inhibition of innate and adaptive immune cell function and infiltration, and enrichment of immunosuppressive cells or molecules [reviewed in (Sharma et al. 2017; Jenkins et al. 2018)].
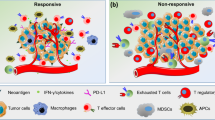
Immune cell profiling can reveal how tumor intrinsic and extrinsic mechanisms hinder anti-tumor responses and contribute to immune checkpoint therapy resistance. A valuable approach to uncover resistance mechanisms involves immune profiling of longitudinal samples. Comparison of the immune contexture in tissues and blood before and during immune checkpoint therapy can provide information on the dynamics of the immune response and expose defective or abnormal immune activity prior to and throughout the course of therapy (Fig. 2).
Defective IFNγ responses
The IFNγ signaling pathway acts as a double-edged sword, with critical roles in mediating the degree and extent of anti-tumor immune response, and by extension, the efficacy of immune checkpoint therapy. For example, IFNγ regulates CD8 T cell expansion but can also induce adaptive immune resistance mechanisms. Alterations in IFNγ pathway components that cause dysfunctional or defective signaling are often associated with resistance to immune checkpoint inhibitors. For instance, loss-of-function mutations in the PBRM1 gene, which encodes a subunit of the PBAF SWI/SNF chromatin remodeling complex, was enriched in tumors of metastatic renal cell carcinoma patients responding to anti-PD-1 therapy. PBRM1/PBAF deficient tumors showed enhanced expression of IL-6 JAK-STAT3 signaling, suggesting that inhibition of this complex may potentiate IFNγ-dependent immune stimulation (Miao et al. 2018). In line with this, Pan et al. identified inactivation of PBRM1, ARID2, and BRD7, components of the PBAF SWI/SNF complex, to render tumor cells sensitive to IFNγ and T cell-mediated killing. Flow cytometric analysis of PBRM1-deficient tumors showed increased infiltration of CD4 T and CD8 T cells, and single-cell RNA sequencing analysis of the immune cells indicated increased expression of immune activation pathways. Importantly, PBRM1 deletion caused treatment resistant tumors to become responsive to immune checkpoint inhibitors (Pan et al. 2018).
Deletion of a protein tyrosine phosphatase PTPN2 has also been shown to sensitize tumors to immune checkpoint inhibitors, through enhanced IFNγ signaling (Manguso et al. 2017). Similarly, loss of function mutations in APLNR gene was identified in non-responding tumor lesions of metastatic melanoma and lung cancer patients treated with anti-PD-1 or anti-CTLA-4 therapy. APLNR was shown to bind JAK1 and induce IFNγ signaling, and its deletion reduced the efficacy of immune checkpoint blockade (Patel et al. 2017). Collectively, these studies utilize immune profiling to demonstrate how dysregulated IFNγ signaling shapes the immune microenvironment and contributes to resistance to immune checkpoint therapy.
T cell exclusion
Alteration in certain oncogenic signaling pathways has been shown to contribute to T cell exclusion, and hence, proposed as a tumor intrinsic resistance mechanism. PTEN loss in cancer cells was shown to reduce accumulation of adoptively transferred T cells and suppress anti-tumor T cell killing in a mouse model. Loss of PTEN, which leads to activation of the PI3K signaling pathway, was also associated with significantly reduced CD8 T cell infiltration in tumor biopsies of metastatic melanoma patients (Peng et al. 2016). The B-catenin/WNT signaling pathway in cancer cells was similarly implicated in T cell exclusion. Activation of WNT signaling suppressed CCL4 expression, leading to reduced recruitment of CD103+ dendritic cells (Spranger et al. 2015), an antigen-presenting immune cell subset essential for effector T cell trafficking into the tumor microenvironment (Spranger et al. 2017).
Inhibition of immune cell activity
Immune cell profiling using flow or mass cytometry provides comprehensive characterization of immune cell phenotype, function, and activity, and can elucidate response and resistance mechanisms of immune checkpoint therapy. For instance, mass cytometry profiling of tumor-infiltrating T cells revealed differences in the T cell phenotypes of patients treated with anti-PD-1 compared to anti-CTLA-4. Treatment with anti-PD-1 appeared to invigorate exhausted CD8 T cells while anti-CTLA-4 treatment induced expression of Th1 CD4 T cells that recruit and activate other effector cytotoxic cells, indicating distinct response mechanisms in the two checkpoint inhibitors (Wei et al. 2017).
On the other hand, resistance to immune checkpoint blockade appears to depend on the balance between T cell activity and its inhibition. Profiling of tumor-infiltrating CD8 T cells in NSCLC patients (n = 32) revealed increased expression of immune inhibitory molecules PD-1, TIM-3, CTLA-4, LAG-3, and BTLA correlating with T cell dysfunction and tumor progression (Thommen et al. 2015), and expression of these immune inhibitory checkpoints may exert an immunosuppressive state despite anti-PD-1 treatment. Profiling of peripheral blood from melanoma patients before and after treatment with anti-PD-1 identified an exhausted CD8 T cell phenotype, which can be reinvigorated with treatment. Interestingly, resistance to anti-PD-1 was not due to a failure in reinvigoration, but rather the magnitude of the reinvigoration in relation to tumor burden (Huang et al. 2017). Exhausted CD8 T cells show sustained levels of PD-1 and a distinct epigenetic landscape compared to other subsets of effector and memory CD8 T cells (Sen et al. 2016); difference in the epigenetic modulation of this exhausted T cell phenotype affects reinvigoration with anti-PD-1 therapy (Pauken et al. 2016), and their ability to be reprogrammed to avoid exhaustion and dysfunction (Philip et al. 2017).
Presence of immunosuppressive cells
Immune cells such as the T regulatory and Th2 T cells, myeloid-derived suppressor cells and M2 polarized and tumor-associated macrophages secrete various anti-inflammatory and immune suppressive factors to dampen T cell-mediated responses. Hence, identification of these cells in the tumor microenvironment using various immune profiling techniques has shed light on why some patients fail immune checkpoint therapy. Indeed, depletion of T regulatory cells has been shown to improve anti-tumor immune response (Viehl et al. 2006), while presence of myeloid-derived suppressor cells correlated with decreased anti-CTLA-4 response (Meyer et al. 2014). Intra-tumoral macrophages were identified in tumors resistant to immune checkpoint blockade in a mouse breast carcinoma model, and targeting of these cells using a selective PI3K inhibitor increased anti-tumor T cell activity (De Henau et al. 2016). Tumor-associated macrophages were also shown to scavenge anti-PD-1, and the removal of PD-1 antibodies from CD8 T cells may impede treatment efficacy (Arlauckas et al. 2017). Targeting of tumor-associated macrophages with anti-CSF-1R reduced their infiltration, and when used in combination with anti-PD-1 or anti-CTLA-4, enhanced tumor regression (Zhu et al. 2014).
Conclusions
The profile and distribution of immune cells play essential roles in determining response to immune checkpoint blockade. Characterization of the immune microenvironment has shown enormous value in identifying prognostic and predictive immune-associated biomarkers that can be implemented in the clinic for patient care. Moreover, immune profiling of resistant tumor biopsies has elucidated the intricate biological processes contributing to failure to immune checkpoint inhibitors, and uncovering these resistance mechanisms will reveal strategies to improve the efficacy of these therapies.
References
Amirel AD, Davis KL, Tadmor MD, Simonds EF, Levine JH, Bendall SC, Shenfeld DK, Krishnaswamy S, Nolan GP, Pe’er D (2013) viSNE enables visualization of high dimensional single-cell data and reveals phenotypic heterogeneity of leukemia. Nat Biotechnol 31: 545–552
Andersen RS, Kvistborg P, Frosig TM, Pedersen NW, Lyngaa R, Bakker AH, Shu CJ, Straten P, Schumacher TN, Hadrup SR (2012) Parallel detection of antigen-specific T cell responses by combinatorial encoding of MHC multimers. Nat Protoc 7:891–902
Arlauckas SP, Garris CS, Kohler RH, Kitaoka M, Cuccarese MF, Yang KS, Miller MA, Carlson JC, Freeman GJ, Anthony RM, Weissleder R, Pittet MJ (2017) In vivo imaging reveals a tumor-associated macrophage-mediated resistance pathway in anti-PD-1 therapy. Sci Transl Med. https://doi.org/10.1126/scitranslmed.aal3604
Ascierto PA, Capone M, Urba WJ, Bifulco CB, Botti G, Lugli A, Marincola FM, Ciliberto G, Galon J, Fox BA (2013) The additional facet of immunoscore: immunoprofiling as a possible predictive tool for cancer treatment. J Transl Med 11:54
Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, Piha-Paul SA, Yearley J, Seiwert TY, Ribas A, McClanahan TK (2017) IFN-gamma-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest 127:2930–2940
Baitsch L, Baumgaertner P, Devevre E, Raghav SK, Legat A, Barba L, Wieckowski S, Bouzourene H, Deplancke B, Romero P, Rufer N, Speiser DE (2011) Exhaustion of tumor-specific CD8(+) T cells in metastases from melanoma patients. J Clin Invest 121:2350–2360
Becht E, Giraldo NA, Dieu-Nosjean MC, Sautes-Fridman C, Fridman WH (2016a) Cancer immune contexture and immunotherapy. Curr Opin Immunol 39:7–13
Becht E, Giraldo NA, Germain C, de Reynies A, Laurent-Puig P, Zucman-Rossi J, Dieu-Nosjean MC, Sautes-Fridman C, Fridman WH (2016b) Immune contexture, immunoscore, and malignant cell molecular subgroups for prognostic and theranostic classifications of cancers. Adv Immunol 130:95–190
Becker M, De Bastiani MA, Parisi MM, Guma FT, Markoski MM, Castro MA, Kaplan MH, Barbe-Tuana FM, Klamt F (2015) Integrated transcriptomics establish macrophage polarization signatures and have potential applications for clinical health and disease. Sci Rep 5:13351
Bendall SC, Nolan GP, Roederer M, Chattopadhyay PK (2012) A deep profiler’s guide to cytometry. Trends Immunol 33:323–332
Bhatia S, Tykodi SS, Thompson JA (2009) Treatment of metastatic melanoma: an overview. Oncology 23:488–496
Bjornson ZB, Nolan GP, Fantl WJ (2013) Single-cell mass cytometry for analysis of immune system functional states. Curr Opin Immunol 25:484–494
Blumenreich MS (1990). The white blood cell and differential count. In: Walker HK, Hall WD, Hurst JW (eds) Clinical methods: the history, physical, and laboratory examinations. Butterworths, Boston
Bogunovic D, O’Neill DW, Belitskaya-Levy I, Vacic V, Yu YL, Adams S, Darvishian F, Berman R, Shapiro R, Pavlick AC, Lonardi S, Zavadil J, Osman I, Bhardwaj N (2009) Immune profile and mitotic index of metastatic melanoma lesions enhance clinical staging in predicting patient survival. Proc Natl Acad Sci USA 106:20429–20434
Chen DS, Mellman I (2013) Oncology meets immunology: the cancer-immunity cycle. Immunity 39:1–10
Chen DS, Mellman I (2017) Elements of cancer immunity and the cancer-immune set point. Nature 541:321–330
Chen PL, Roh W, Reuben A, Cooper ZA, Spencer CN, Prieto PA, Miller JP, Bassett RL, Gopalakrishnan V, Wani K, De Macedo MP, Austin-Breneman JL, Jiang H, Chang Q, Reddy SM, Chen WS, Tetzlaff MT, Broaddus RJ, Davies MA, Gershenwald JE, Haydu L, Lazar AJ, Patel SP, Hwu P, Hwu WJ, Diab A, Glitza IC, Woodman SE, Vence LM, Wistuba II, Amaria RN, Kwong LN, Prieto V, Davis RE, Ma W, Overwijk WW, Sharpe AH, Hu J, Futreal PA, Blando J, Sharma P, Allison JP, Chin L, Wargo JA (2016) Analysis of immune signatures in longitudinal tumor samples yields insight into biomarkers of response and mechanisms of resistance to immune checkpoint blockade. Cancer Discov 6:827–837
Covey TM, Cesano A (2010) Modulated multiparametric phosphoflow cytometry in hematological malignancies: technology and clinical applications. Best Pract Res Clin Haematol 23:319–331
Critchley-Thorne RJ, Yu HX, Lee PP (2011) Immune signatures associated with the cancer bearing state. In: Immunologic signatures of rejection. Springer, New York, pp. 169–186
Curiel TJ, Coukos G, Zou L, Alvarez X, Cheng P, Mottram P, Evdemon-Hogan M, Conejo-Garcia JR, Zhang L, Burow M, Zhu Y, Wei S, Kryczek I, Daniel B, Gordon A, Myers L, Lackner A, Disis ML, Knutson KL, Chen L, Zou W (2004) Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat Med 10:942–949
Daud A, Ribas A, Robert C, Hodi FS, Wolchok JD, Joshua AM, Hwu WJ, Weber JS, Gangadhar TC, Joseph RW, Dronca RS, Patnaik A, Zarour HM, Kefford R, Lindia JA, Li XN, Ebbinghaus S, Kang SP, Hamid O (2015) Long-term efficacy of pembrolizumab (pembro; MK-3475) in a pooled analysis of 655 patients (pts) with advanced melanoma (MEL) enrolled in KEYNOTE-001. J Clin Oncol 33
Daud AI, Loo K, Pauli ML, Sanchez-Rodriguez R, Sandoval PM, Taravati K, Tsai K, Nosrati A, Nardo L, Alvarado MD, Algazi AP, Pampaloni MH, Lobach IV, Hwang J, Pierce RH, Gratz IK, Krummel MF, Rosenblum MD (2016) Tumor immune profiling predicts response to anti-PD-1 therapy in human melanoma. J Clin Invest 126:3447–3452
De Matos LL, Trufelli DC, de Matos MGL, da Silva Pinhal MA (2010) Immunohistochemistry as an important tool in biomarkers detection and clinical practice. Biomarker Insights 5:9–20
De Henau O, Rausch M, Winkler D, Campesato LF, Liu C, Cymerman DH, Budhu S, Ghosh A, Pink M, Tchaicha J, Douglas M, Tibbitts T, Sharma S, Proctor J, Kosmider N, White K, Stern H, Soglia J, Adams J, Palombella VJ, McGovern K, Kutok JL, Wolchok JD, Merghoub T (2016) Overcoming resistance to checkpoint blockade therapy by targeting PI3Kgamma in myeloid cells. Nature 539:443–447
Dixon AR, Bathany C, Tsuei M, White J, Barald KF, Takayama S (2015) Recent developments in multiplexing techniques for immunohistochemistry. Expert Rev Mol Diagn 15:1171–1186
Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D, Artal-Cortes A, Lewanski C, Braiteh F, Waterkamp D, He P, Zou W, Chen DS, Yi J, Sandler A, Rittmeyer A, Group PS (2016) Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet 387:1837–1846
Finn OJ (2003) Cancer vaccines: between the idea and the reality. Nat Rev Immunol 3:630–641
Finotello F, Trajanoski Z (2018) Quantifying tumor-infiltrating immune cells from transcriptomics data. Cancer Immunol Immunother 1–10
Fridman WH, Pages F, Sautes-Fridman C, Galon J (2012) The immune contexture in human tumors: impact on clinical outcome. Nat Rev Cancer 12:298–306
Fridman WH, Zitvogel L, Sautes-Fridman C, Kroemer G (2017) The immune contexture in cancer prognosis and treatment. Nat Rev Clin Oncol 14:717–734
Gajewski TF, Louahed J, Brichard VG (2010) Gene signature in melanoma associated with clinical activity: a potential clue to unlock cancer immunotherapy. Cancer J 16:399–403
Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pages C, Tosolini M, Camus M, Berger A, Wind P, Zinzindohoue F, Bruneval P, Cugnenc PH, Trajanoski Z, Fridman WH, Pages F (2006) Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science 313:1960–1964
Galon J, Pages F, Marincola FM, Angell HK, Thurin M, Lugli A, Zlobec I, Berger A, Bifulco C, Botti G, Tatangelo F, Britten CM, Kreiter S, Chouchane L, Delrio P, Arndt H, Asslaber M, Maio M, Masucci GV, Mihm M, Vidal-Vanaclocha F, Allison JP, Gnjatic S, Hakansson L, Huber C, Singh-Jasuja H, Ottensmeier C, Zwierzina H, Laghi L, Grizzi F, Ohashi PS, Shaw PA, Clarke BA, Wouters BG, Kawakami Y, Hazama S, Okuno K, Wang E, O’Donnell-Tormey J, Lagorce C, Pawelec G, Nishimura MI, Hawkins R, Lapointe R, Lundqvist A, Khleif SN, Ogino S, Gibbs P, Waring P, Sato N, Torigoe T, Itoh K, Patel PS, Shukla SN, Palmqvist R, Nagtegaal ID, Wang Y, D’Arrigo C, Kopetz S, Sinicrope FA, Trinchieri G, Gajewski TF, Ascierto PA, Fox BA (2012). Cancer classification using the Immunoscore: a worldwide task force. J Transl Med 10:205
Galon J, Mlecnik B, Bindea G, Angell HK, Berger A, Lagorce C, Lugli A, Zlobec I, Hartmann A, Bifulco C, Nagtegaal ID, Palmqvist R, Masucci GV, Botti G, Tatangelo F, Delrio P, Maio M, Laghi L, Grizzi F, Asslaber M, D’Arrigo C, Vidal-Vanaclocha F, Zavadova E, Chouchane L, Ohashi PS, Hafezi-Bakhtiari S, Wouters BG, Roehrl M, Nguyen L, Kawakami Y, Hazama S, Okuno K, Ogino S, Gibbs P, Waring P, Sato N, Torigoe T, Itoh K, Patel PS, Shukla SN, Wang Y, Kopetz S, Sinicrope FA, Scripcariu V, Ascierto PA, Marincola FM, Fox BA, Pages F (2014) Towards the introduction of the ‘Immunoscore’ in the classification of malignant tumors. J Pathol 232:199–209
Garcia-Diaz A, Shin DS, Moreno BH, Saco J, Escuin-Ordinas H, Rodriguez GA, Zaretsky JM, Sun L, Hugo W, Wang X, Parisi G, Saus CP, Torrejon DY, Graeber TG, Comin-Anduix B, Hu-Lieskovan S, Damoiseaux R, Lo RS, Ribas A (2017) Interferon receptor signaling pathways regulating PD-L1 and PD-L2 expression. Cell Rep 19:1189–1201
Godec J, Tan Y, Liberzon A, Tamayo P, Bhattacharya S, Butte AJ, Mesirov JP, Haining WN (2016) Compendium of immune signatures identifies conserved and species-specific biology in response to inflammation. Immunity 44:194–206
Hamid O, Schmidt H, Nissan A, Ridolfi L, Aamdal S, Hansson J, Guida M, Hyams DM, Gomez H, Bastholt L, Chasalow SD, Berman D (2011) A prospective phase II trial exploring the association between tumor microenvironment biomarkers and clinical activity of ipilimumab in advanced melanoma. J Transl Med 9:204
Heng TS, Painter MW, Immunological Genome Project C (2008) The immunological genome project: networks of gene expression in immune cells. Nat Immunol 9:1091–1094
Hodi FS, Chesney J, Pavlick AC, Robert C, Grossmann KF, McDermott DF, Linette GP, Meyer N, Giguere JK, Agarwala SS, Shaheen M, Ernstoff MS, Minor DR, Salama AK, Taylor MH, Ott PA, Horak C, Gagnier P, Jiang J, Wolchok JD, Postow MA (2016) Combined nivolumab and ipilimumab versus ipilimumab alone in patients with advanced melanoma: 2-year overall survival outcomes in a multicentre, randomised, controlled, phase 2 trial. Lancet Oncol 17:1558–1568
Huang AC, Postow MA, Orlowski RJ, Mick R, Bengsch B, Manne S, Xu W, Harmon S, Giles JR, Wenz B, Adamow M, Kuk D, Panageas KS, Carrera C, Wong P, Quagliarello F, Wubbenhorst B, D’Andrea K, Pauken KE, Herati RS, Staupe RP, Schenkel JM, McGettigan S, Kothari S, George SM, Vonderheide RH, Amaravadi RK, Karakousis GC, Schuchter LM, Xu X, Nathanson KL, Wolchok JD, Gangadhar TC, Wherry EJ (2017) T-cell invigoration to tumor burden ratio associated with anti-PD-1 response. Nature 545:60–65
Hugo W, Zaretsky JM, Sun L, Song C, Moreno BH, Hu-Lieskovan S, Berent-Maoz B, Pang J, Chmielowski B, Cherry G, Seja E, Lomeli S, Kong X, Kelley MC, Sosman JA, Johnson DB, Ribas A, Lo RS (2017) Genomic and transcriptomic features of response to Anti-PD-1 therapy in metastatic melanoma. Cell 168:542
Jenkins RW, Barbie DA, Flaherty KT (2018) Mechanisms of resistance to immune checkpoint inhibitors. Br J Cancer 118:9–16
Ji RR, Chasalow SD, Wang L, Hamid O, Schmidt H, Cogswell J, Alaparthy S, Berman D, Jure-Kunkel M, Siemers NO, Jackson JR, Shahabi V (2012) An immune-active tumor microenvironment favors clinical response to ipilimumab. Cancer Immunol Immunother 61:1019–1031
Kantoff PW, Higano CS, Shore ND, Berger ER, Small EJ, Penson DF, Redfern CH, Ferrari AC, Dreicer R, Sims RB, Xu Y, Frohlich MW, Schellhammer PF, Investigators IS (2010) Sipuleucel-T immunotherapy for castration-resistant prostate cancer. N Engl J Med 363:411–422
Karachaliou N, Gonzalez-Cao M, Crespo G, Drozdowskyj A, Aldeguer E, Gimenez-Capitan A, Teixido C, Molina-Vila MA, Viteri S, De Los Llanos Gil M, Algarra SM, Perez-Ruiz E, Marquez-Rodas I, Rodriguez-Abreu D, Blanco R, Puertolas T, Royo MA, Rosell R (2018) Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol 10:1758834017749748
Krieg C, Nowicka M, Guglietta S, Schindler S, Hartmann FJ, Weber LM, Dummer R, Robinson MD, Levesque MP, Becher B (2018) High-dimensional single-cell analysis predicts response to anti-PD-1 immunotherapy. Nat Med 24:144–153
Krutzik PO, Irish JM, Nolan GP, Perez OD (2004) Analysis of protein phosphorylation and cellular signaling events by flow cytometry: techniques and clinical applications. Clin Immunol 110:206–221
Kulasingam V, Diamandis EP (2008) Strategies for discovering novel cancer biomarkers through utilization of emerging technologies. Nat Clin Pract Oncol 5:588–599
Larkin J, Hodi FS, Wolchok JD (2015) Combined nivolumab and Ipilimumab or monotherapy in untreated melanoma. N Engl J Med 373:1270–1271
Li B, Severson E, Pignon JC, Zhao H, Li T, Novak J, Jiang P, Shen H, Aster JC, Rodig S, Signoretti S, Liu JS, Liu XS (2016) Comprehensive analyses of tumor immunity: implications for cancer immunotherapy. Genome Biol 17:174
Liu C, Peng W, Xu C, Lou Y, Zhang M, Wargo JA, Chen JQ, Li HS, Watowich SS, Yang Y, Frederick DT, Cooper ZA, Mbofung RM, Whittington M, Flaherty KT, Woodman SE, Davies MA, Radvanyi LG, Overwijk WW, Lizee G, Hwu P (2013) BRAF inhibition increases tumor infiltration by T cells and enhances the antitumor activity of adoptive immunotherapy in mice. Clin Cancer Res 19:393–403
Maecker HT, McCoy JP, Nussenblatt R (2012) Standardizing immunophenotyping for the human immunology project. Nat Rev Immunol 12:191–200
Mandruzzato S, Callegaro A, Turcatel G, Francescato S, Montesco MC, Chiarion-Sileni V, Mocellin S, Rossi CR, Bicciato S, Wang E, Marincola FM, Zanovello P (2006) A gene expression signature associated with survival in metastatic melanoma. J Transl Med 4:50
Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, Collins NB, Bi K, LaFleur MW, Juneja VR, Weiss SA, Lo J, Fisher DE, Miao D, Van Allen E, Root DE, Sharpe AH, Doench JG, Haining WN (2017) In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature 547:413–418
Martens A, Wistuba-Hamprecht K, Foppen MG, Yuan J, Postow MA, Wong P, Romano E, Khammari A, Dreno B, Capone M, Ascierto PA, Di Giacomo AM, Maio M, Schilling B, Sucker A, Schadendorf D, Hassel JC, Eigentler TK, Martus P, Wolchok JD, Blank C, Pawelec G, Garbe C, Weide B (2016) Baseline peripheral blood biomarkers associated with clinical outcome of advanced melanoma patients treated with ipilimumab. Clin Cancer Res 22:2908–2918
McDermott D, Lebbe C, Hodi FS, Maio M, Weber JS, Wolchok JD, Thompson JA, Balch CM (2014) Durable benefit and the potential for long-term survival with immunotherapy in advanced melanoma. Cancer Treat Rev 40:1056–1064
McDermott DF, Sosman JA, Sznol M, Massard C, Gordon MS, Hamid O, Powderly JD, Infante JR, Fasso M, Wang YV, Zou W, Hegde PS, Fine GD, Powles T (2016) Atezolizumab, an anti-programmed death-ligand 1 antibody, in metastatic renal cell carcinoma: long-term safety, clinical activity, and immune correlates from a phase ia study. J Clin Oncol 34:833–842
Melief CJ, van Hall T, Arens R, Ossendorp F, van der Burg SH (2015) Therapeutic cancer vaccines. J Clin Invest 125:3401–3412
Meyer C, Cagnon L, Costa-Nunes CM, Baumgaertner P, Montandon N, Leyvraz L, Michielin O, Romano E, Speiser DE (2014) Frequencies of circulating MDSC correlate with clinical outcome of melanoma patients treated with ipilimumab. Cancer Immunol Immunother 63:247–257
Mezquita L, Charrier M, Auclin E, Gion M, Remon J, Planchard D, Dupraz L, Lahmar J, Gazzah A, Chaput N, Besse B (2017) Neutrophil-To-lymphocyte and other ratios as prognostic and predictive markers of immune checkpoint inhibitors in advanced NSCLC patients. J Thorac Oncol 12:S1315–S1316
Miao D, Margolis CA, Gao W, Voss MH, Li W, Martini DJ, Norton C, Bosse D, Wankowicz SM, Cullen D, Horak C, Wind-Rotolo M, Tracy A, Giannakis M, Hodi FS, Drake CG, Ball MW, Allaf ME, Snyder A, Hellmann MD, Ho T, Motzer RJ, Signoretti S, Kaelin WG Jr, Choueiri TK, Van Allen EM (2018) Genomic correlates of response to immune checkpoint therapies in clear cell renal cell carcinoma. Science 359:801–806
Muenst S, Laubli H, Soysal SD, Zippelius A, Tzankov A, Hoeller S (2016) The immune system and cancer evasion strategies: therapeutic concepts. J Intern Med 279:541–562
Newell EW (2013) Higher throughput methods of identifying T cell epitopes for studying outcomes of altered antigen processing and presentation. Front Immunol 4:430
Newick K, O’Brien S, Moon E, Albelda SM (2017) CAR T cell therapy for solid tumors. Annu Rev Med 68:139–152
Newman AM, Liu CL, Green MR, Gentles AJ, Feng W, Xu Y, Hoang CD, Diehn M, Alizadeh AA (2015) Robust enumeration of cell subsets from tissue expression profiles. Nat Methods 12:453–457
Ng Tang D, Shen Y, Sun J, Wen S, Wolchok JD, Yuan J, Allison JP, Sharma P (2013) Increased frequency of ICOS + CD4 T cells as a pharmacodynamic biomarker for anti-CTLA-4 therapy. Cancer Immunol Res 1:229–234
Ornatsky O, Bandura D, Baranov V, Nitz M, Winnik MA, Tanner S (2010) Highly multiparametric analysis by mass cytometry. J Immunol Methods 361:1–20
Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, Tsoucas D, Qiu X, Lim K, Rao P, Long HW, Yuan GC, Doench J, Brown M, Liu XS, Wucherpfennig KW (2018) A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 359:770–775
Pardoll DM (2012) The blockade of immune checkpoints in cancer immunotherapy. Nat Rev Cancer 12:252–264
Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, Gartner JJ, Jia L, Steinberg SM, Yamamoto TN, Merchant AS, Mehta GU, Chichura A, Shalem O, Tran E, Eil R, Sukumar M, Guijarro EP, Day CP, Robbins P, Feldman S, Merlino G, Zhang F, Restifo NP (2017) Identification of essential genes for cancer immunotherapy. Nature 548(7669):537
Pauken KE, Sammons MA, Odorizzi PM, Manne S, Godec J, Khan O, Drake AM, Chen Z, Sen DR, Kurachi M, Barnitz RA, Bartman C, Bengsch B, Huang AC, Schenkel JM, Vahedi G, Haining WN, Berger SL, Wherry EJ (2016) Epigenetic stability of exhausted T cells limits durability of reinvigoration by PD-1 blockade. Science 354:1160–1165
Peng W, Chen JQ, Liu C, Malu S, Creasy C, Tetzlaff MT, Xu C, McKenzie JA, Zhang C, Liang X, Williams LJ, Deng W, Chen G, Mbofung R, Lazar AJ, Torres-Cabala CA, Cooper ZA, Chen PL, Tieu TN, Spranger S, Yu X, Bernatchez C, Forget MA, Haymaker C, Amaria R, McQuade JL, Glitza IC, Cascone T, Li HS, Kwong LN, Heffernan TP, Hu J, Bassett RL Jr, Bosenberg MW, Woodman SE, Overwijk WW, Lizee G, Roszik J, Gajewski TF, Wargo JA, Gershenwald JE, Radvanyi L, Davies MA, Hwu P (2016) Loss of PTEN promotes resistance to T cell-mediated immunotherapy. Cancer Discov 6:202–216
Perfetto SP, Chattopadhyay PK, Roederer M (2004) Seventeen-colour flow cytometry: unravelling the immune system. Nat Rev Immunol 4:648–655
Philip M, Fairchild L, Sun L, Horste EL, Camara S, Shakiba M, Scott AC, Viale A, Lauer P, Merghoub T, Hellmann MD, Wolchok JD, Leslie CS, Schietinger A (2017) Chromatin states define tumor-specific T cell dysfunction and reprogramming. Nature 545:452–456
Postow MA, Manuel M, Wong P, Yuan J, Dong Z, Liu C, Perez S, Tanneau I, Noel M, Courtier A, Pasqual N, Wolchok JD (2015) Peripheral T cell receptor diversity is associated with clinical outcomes following ipilimumab treatment in metastatic melanoma. J Immunother Cancer 3:23
Qiu P, Simonds EF, Bendall SC, Gibbs KD Jr, Bruggner RV, Linderman MD, Sachs K, Nolan GP, Plevritis SK (2011) Extracting a cellular hierarchy from high-dimensional cytometry data with SPADE. Nat Biotechnol 29:886–891
Ramos CA, Heslop HE, Brenner MK (2016) CAR-T cell therapy for lymphoma. Annu Rev Med 67:165–183
Restifo NP, Dudley ME, Rosenberg SA (2012) Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol 12:269–281
Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martin-Algarra S, Mandal R, Sharfman WH, Bhatia S, Hwu WJ, Gajewski TF, Slingluff CL Jr, Chowell D, Kendall SM, Chang H, Shah R, Kuo F, Morris LGT, Sidhom JW, Schneck JP, Horak CE, Weinhold N, Chan TA (2017) Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell 171:934–949
Ribas A, Hamid O, Daud A, Hodi FS, Wolchok JD, Kefford R, Joshua AM, Patnaik A, Hwu WJ, Weber JS, Gangadhar TC, Hersey P, Dronca R, Joseph RW, Zarour H, Chmielowski B, Lawrence DP, Algazi A, Rizvi NA, Hoffner B, Mateus C, Gergich K, Lindia JA, Giannotti M, Li XN, Ebbinghaus S, Kang SP, Robert C (2016) Association of pembrolizumab with tumor response and survival among patients with advanced melanoma. JAMA 315:1600–1609
Robert C, Schachter J, Long GV, Arance A, Grob JJ, Mortier L, Daud A, Carlino MS, McNeil C, Lotem M, Larkin J, Lorigan P, Neyns B, Blank CU, Hamid O, Mateus C, Shapira-Frommer R, Kosh M, Zhou H, Ibrahim N, Ebbinghaus S, Ribas A (2015) Pembrolizumab versus ipilimumab in advanced melanoma. N Engl J Med 372:2521–2532
Roepman P, Jassem J, Smit EF, Muley T, Niklinski J, van de Velde T, Witteveen AT, Rzyman W, Floore A, Burgers S, Giaccone G, Meister M, Dienemann H, Skrzypski M, Kozlowski M, Mooi WJ, van Zandwijk N (2009) An immune response enriched 72-gene prognostic profile for early-stage non-small-cell lung cancer. Clin Cancer Res 15:284–290
Rooney MS, Shukla SA, Wu CJ, Getz G, Hacohen N (2015) Molecular and genetic properties of tumors associated with local immune cytolytic activity. Cell 160:48–61
Sato E, Olson SH, Ahn J, Bundy B, Nishikawa H, Qian F, Jungbluth AA, Frosina D, Gnjatic S, Ambrosone C, Kepner J, Odunsi T, Ritter G, Lele S, Chen YT, Ohtani H, Old LJ, Odunsi K (2005) Intraepithelial CD8+ tumor-infiltrating lymphocytes and a high CD8+/regulatory T cell ratio are associated with favorable prognosis in ovarian cancer. Proc Natl Acad Sci USA 102:18538–18543
Schadendorf D, Hodi FS, Robert C, Weber JS, Margolin K, Hamid O, Patt D, Chen TT, Berman DM, Wolchok JD (2015) Pooled analysis of long-term survival data from phase II and phase III trials of ipilimumab in unresectable or metastatic melanoma. J Clin Oncol 33:1889–1894
Sen DR, Kaminski J, Barnitz RA, Kurachi M, Gerdemann U, Yates KB, Tsao HW, Godec J, LaFleur MW, Brown FD, Tonnerre P, Chung RT, Tully DC, Allen TM, Frahm N, Lauer GM, Wherry EJ, Yosef N, Haining WN (2016) The epigenetic landscape of T cell exhaustion. Science 354:1165–1169
Shaffer AL, Rosenwald A, Hurt EM, Giltnane JM, Lam LT, Pickeral OK, Staudt LM (2001) Signatures of the immune response. Immunity 15:375–385
Sharma P, Hu-Lieskovan S, Wargo JA, Ribas A (2017) Primary, adaptive, and acquired resistance to cancer immunotherapy. Cell 168:707–723
Shin DS, Zaretsky JM, Escuin-Ordinas H, Garcia-Diaz A, Hu-Lieskovan S, Kalbasi A, Grasso CS, Hugo W, Sandoval S, Torrejon DY, Palaskas N, Rodriguez GA, Parisi G, Azhdam A, Chmielowski B, Cherry G, Seja E, Berent-Maoz B, Shintaku IP, Le DT, Pardoll DM, Diaz LA Jr, Tumeh PC, Graeber TG, Lo RS, Comin-Anduix B, Ribas A (2017) Primary resistance to PD-1 blockade mediated by JAK1/2 mutations. Cancer Discov 7:188–201
Spitzer MH, Nolan GP (2016) Mass cytometry: single cells, many features. Cell 165:780–791
Spranger S, Bao R, Gajewski TF (2015) Melanoma-intrinsic beta-catenin signalling prevents anti-tumor immunity. Nature 523:231–235
Spranger S, Dai D, Horton B, Gajewski TF (2017) Tumor-residing Batf3 dendritic cells are required for effector T cell trafficking and adoptive T cell therapy. Cancer Cell 31:711–723
Stack EC, Wang C, Roman KA, Hoyt CC (2014) Multiplexed immunohistochemistry, imaging, and quantitation: a review, with an assessment of Tyramide signal amplification, multispectral imaging and multiplex analysis. Methods 70:46–58
Subrahmanyam PB, Dong Z, Gusenleitner D, Giobbie-Hurder A, Severgnini M, Zhou J, Manos M, Eastman LM, Maecker HT, Hodi FS (2018) Distinct predictive biomarker candidates for response to anti-CTLA-4 and anti-PD-1 immunotherapy in melanoma patients. J Immunother Cancer 6:18
Thommen DS, Schreiner J, Muller P, Herzig P, Roller A, Belousov A, Umana P, Pisa P, Klein C, Bacac M, Fischer OS, Moersig W, Savic Prince S, Levitsky V, Karanikas V, Lardinois D, Zippelius A (2015) Progression of lung cancer is associated with increased dysfunction of T cells defined by coexpression of multiple inhibitory receptors. Cancer Immunol Res 3:1344–1355
Tirosh I, Izar B, Prakadan SM, Wadsworth MH 2nd, Treacy D, Trombetta JJ, Rotem A, Rodman C, Lian C, Murphy G, Fallahi-Sichani M, Dutton-Regester K, Lin JR, Cohen O, Shah P, Lu D, Genshaft AS, Hughes TK, Ziegler CG, Kazer SW, Gaillard A, Kolb KE, Villani AC, Johannessen CM, Andreev AY, Van Allen EM, Bertagnolli M, Sorger PK, Sullivan RJ, Flaherty KT, Frederick DT, Jane-Valbuena J, Yoon CH, Rozenblatt-Rosen O, Shalek AK, Regev A, Garraway LA (2016) Dissecting the multicellular ecosystem of metastatic melanoma by single-cell RNA-sEq. Science 352:189–196
Topalian SL, Drake CG, Pardoll DM (2015) Immune checkpoint blockade: a common denominator approach to cancer therapy. Cancer Cell 27:450–461
Tumeh PC, Harview CL, Yearley JH, Shintaku IP, Taylor EJ, Robert L, Chmielowski B, Spasic M, Henry G, Ciobanu V, West AN, Carmona M, Kivork C, Seja E, Cherry G, Gutierrez AJ, Grogan TR, Mateus C, Tomasic G, Glaspy JA, Emerson RO, Robins H, Pierce RH, Elashoff DA, Robert C, Ribas A (2014) PD-1 blockade induces responses by inhibiting adaptive immune resistance. Nature 515:568–571
Ulloa-Montoya F, Louahed J, Dizier B, Gruselle O, Spiessens B, Lehmann FF, Suciu S, Kruit WH, Eggermont AM, Vansteenkiste J, Brichard VG (2013) Predictive gene signature in MAGE-A3 antigen-specific cancer immunotherapy. J Clin Oncol 31:2388–2395
van Dongen JJ, Lhermitte L, Bottcher S, Almeida J, van der Velden VH, Flores-Montero J, Rawstron A, Asnafi V, Lecrevisse Q, Lucio P, Mejstrikova E, Szczepanski T, Kalina T, de Tute R, Bruggemann M, Sedek L, Cullen M, Langerak AW, Mendonca A, Macintyre E, Martin-Ayuso M, Hrusak O, Vidriales MB, Orfao A, EuroFlow C (2012) EuroFlow antibody panels for standardized n-dimensional flow cytometric immunophenotyping of normal, reactive and malignant leukocytes. Leukemia 26:1908–1975
Van Allen EM, Miao D, Schilling B, Shukla SA, Blank C, Zimmer L, Sucker A, Hillen U, Foppen MHG, Goldinger SM, Utikal J, Hassel JC, Weide B, Kaehler KC, Loquai C, Mohr P, Gutzmer R, Dummer R, Gabriel S, Wu CJ, Schadendorf D, Garraway LA (2015) Genomic correlates of response to CTLA-4 blockade in metastatic melanoma. Science 350:207–211
van der Burg SH, Arens R, Ossendorp F, van Hall T, Melief CJ (2016) Vaccines for established cancer: overcoming the challenges posed by immune evasion. Nat Rev Cancer 16:219–233
Viehl CT, Moore TT, Liyanage UK, Frey DM, Ehlers JP, Eberlein TJ, Goedegebuure PS, Linehan DC (2006) Depletion of CD4+ CD25+ regulatory T cells promotes a tumor-specific immune response in pancreas cancer-bearing mice. Ann Surg Oncol 13:1252–1258
Vilain RE, Menzies AM, Wilmott JS, Kakavand H, Madore J, Guminski A, Liniker E, Kong BY, Cooper AJ, Howle JR, Saw RPM, Jakrot V, Lo S, Thompson JF, Carlino MS, Kefford RF, Long GV, Scolyer RA (2017) Dynamic changes in PD-L1 expression and immune infiltrates early during treatment predict response to PD-1 blockade in melanoma. Clin Cancer Res 23:5024–5033
Vinay DS, Ryan EP, Pawelec G, Talib WH, Stagg J, Elkord E, Lichtor T, Decker WK, Whelan RL, Kumara H, Signori E, Honoki K, Georgakilas AG, Amin A, Helferich WG, Boosani CS, Guha G, Ciriolo MR, Chen S, Mohammed SI, Azmi AS, Keith WN, Bilsland A, Bhakta D, Halicka D, Fujii H, Aquilano K, Ashraf SS, Nowsheen S, Yang X, Choi BK, Kwon BS (2015) Immune evasion in cancer: mechanistic basis and therapeutic strategies. Semin Cancer Biol 35(Suppl):S185–S198
Wei SC, Levine JH, Cogdill AP, Zhao Y, Anang NAS, Andrews MC, Sharma P, Wang J, Wargo JA, Pe’er D, Allison JP (2017) Distinct cellular mechanisms underlie anti-CTLA-4 and anti-PD-1 checkpoint blockade. Cell 170:1120–1133
Weide B, Martens A, Hassel JC, Berking C, Postow MA, Bisschop K, Simeone E, Mangana J, Schilling B, Di Giacomo AM, Brenner N, Kahler K, Heinzerling L, Gutzmer R, Bender A, Gebhardt C, Romano E, Meier F, Martus P, Maio M, Blank C, Schadendorf D, Dummer R, Ascierto PA, Hospers G, Garbe C, Wolchok JD (2016) Baseline biomarkers for outcome of melanoma patients treated with pembrolizumab. Clin Cancer Res 22:5487–5496
Yang JC, Rosenberg SA (2016) Adoptive T-cell therapy for cancer. Adv Immunol 130:279–294
Zaretsky JM, Garcia-Diaz A, Shin DS, Escuin-Ordinas H, Hugo W, Hu-Lieskovan S, Torrejon DY, Abril-Rodriguez G, Sandoval S, Barthly L, Saco J, Homet Moreno B, Mezzadra R, Chmielowski B, Ruchalski K, Shintaku IP, Sanchez PJ, Puig-Saus C, Cherry G, Seja E, Kong X, Pang J, Berent-Maoz B, Comin-Anduix B, Graeber TG, Tumeh PC, Schumacher TN, Lo RS, Ribas A (2016) Mutations associated with acquired resistance to PD-1 blockade in melanoma. N Engl J Med 375:819–829
Zhang L, Conejo-Garcia JR, Katsaros D, Gimotty PA, Massobrio M, Regnani G, Makrigiannakis A, Gray H, Schlienger K, Liebman MN, Rubin SC, Coukos G (2003) Intratumoral T cells, recurrence, and survival in epithelial ovarian cancer. N Engl J Med 348:203–213
Zhu Y, Knolhoff BL, Meyer MA, Nywening TM, West BL, Luo J, Wang-Gillam A, Goedegebuure SP, Linehan DC, DeNardo DG (2014) CSF1/CSF1R blockade reprograms tumor-infiltrating macrophages and improves response to T-cell checkpoint immunotherapy in pancreatic cancer models. Cancer Res 74:5057–5069
Acknowledgements
SYL is supported by Grant #1123911, awarded through the Priority-driven Collaborative Cancer Research Scheme and co-funded by Cancer Australia and Cure Cancer Australia Foundation. HR is supported by an NHMRC Senior Research Fellowship.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
On behalf of all authors, the corresponding author states that there is no conflict of interest.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Lim, S.Y., Rizos, H. Immune cell profiling in the age of immune checkpoint inhibitors: implications for biomarker discovery and understanding of resistance mechanisms. Mamm Genome 29, 866–878 (2018). https://doi.org/10.1007/s00335-018-9757-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-018-9757-4