Abstract
The availability of and access to quality genetically defined, health-status known mouse resources is critical for biomedical research. By ensuring that mice used in research experiments are biologically, genetically, and health-status equivalent, we enable knowledge transfer, hypothesis building based on multiple data streams, and experimental reproducibility based on common mouse resources (reagents). Major repositories for mouse resources have developed over time and each has significant unique resources to offer. Here we (a) describe The International Mouse Strain Resource that offers users a combined catalog of worldwide mouse resources (live, cryopreserved, embryonic stem cells), with direct access to repository sites holding resources of interest and (b) discuss the commitment to nomenclature standards among resources that remain a challenge in unifying mouse resource catalogs.
Similar content being viewed by others

Avoid common mistakes on your manuscript.
Introduction
The IMSR began as a collaboration between MRC Harwell and the Mouse Genome Database (MGD) at the Jackson Laboratory to provide the research community with a common site for finding mouse resources (Eppig and Strivens 1999). The IMSR has since grown to be a multi-institutional international collaboration supporting the research community that uses mouse as a model system for studying biology and disease (Eppig et al. 2005). The goal of the IMSR remains to provide a web searchable catalog to assist investigators in finding the mouse resources needed for their studies. The IMSR produces a continuously updated global resource for scientific investigators. Collaborating repositories contribute data on the resources held in their individual repositories via FTP files regularly submitted to the IMSR database. These data are processed and integrated with data from other repositories, creating a collective global compendium of resources available worldwide. These integrated resources are then provided on a searchable website (www.findmice.org) with links to each repository, the repository’s ordering form, the repository’s web page describing each holding, and links to the Mouse Genome Database (MGD, www.informatics.jax.org) for information about the gene(s) involved, the specific mutations and their genetic backgrounds, the abnormal phenotypes manifested by the mice, and human disease model assertions based on author-reported experimental data with links to the relevant MGD disease page and to OMIM (Online Mendelian Inheritance in Man) records for human disease descriptions.
The IMSR is a dynamic data system. As repositories submit their current holdings, IMSR performs quality control (QC) checks on the data and, if the data files are properly formed, processes the files and refreshes the web presentation to reflect current repository holdings. Ongoing curation provided by MGD staff feeds back to the repositories to continually improve the standardization of nomenclature among repositories and thus iteratively improving the ability of researchers to effectively query IMSR (and their representative repository sites) using standard official nomenclature and retrieve complete results.
International mouse strain resource (IMSR): integrating repository holdings worldwide
Principles and design
The goal of the IMSR is to provide an online searchable web-based catalog of mouse resources available globally, including inbred, mutant, and genetically engineered mice, cryopreserved embryos and gametes, and ES cell lines. The IMSR website provides, for each strain or cell line, links for ordering, links to the repositories’ strain description, and links to phenotype and disease model data. Mouse repositories of any size and in any location are welcome to contribute data about their mouse resource holdings, providing those holdings are available to investigators who request resource access. This does not mean the resources are without cost, but that they are available to researchers. Most repositories charge customers to recover their cost of operating, and maintaining and shipping mouse resources. In addition, IMSR expects that resources will update their holdings on a regular basis. Many active repositories provide new data files on a weekly basis. Individual investigators are welcome to contribute to IMSR as a small repository as well, if they are distributing their resources and will ship their unique mouse mutants without special restrictions.
Repositories contributing to IMSR
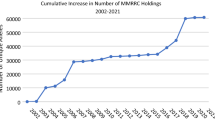
There are currently 20 repositories and repository consortia (representing 46 individual repository sites) listing mouse resource holdings in IMSR (Table 1). These collectively hold 32,396 mouse strains (as live stocks, cryopreserved embryos, and cryopreserved gametes) and 209,328 mutant ES cell lines as of May 15, 2015 (Table 1). Of these, approximately 1300 strains exist as both ES cell lines and as some animated form (largely as cryopreserved sperm or embryos). There is virtually no duplication in strain holdings between repositories. However, at any given time, a repository may have available multiple forms of a given strain (e.g., frozen embryos or live mice) either through the dynamic cycle of cryopreservation, recovery, breeding, and re-cryopreservation that happens in providing or restoring a given repository’s strain holdings or as a matter of repository policy to store strains in multiple states (e.g., as cryopreserved embryos and sperm).
Large-scale mutagenesis and analysis projects including the International Mouse Phenotyping Consortium (IMPC) mice recovered from ES cell line knockout mutations and mice generated in subsequent crosses to cre-deleter mice for removal of neo-cassettes (Brown and Moore 2012; Mallon et al. 2012) and several N-ethyl-N-nitrosourea (ENU) targeted phenotype screens (Li et al. 2015; Arnold et al. 2012; Goldowitz et al. 2004; Nolan et al. 2000; Hrabé de Angelis et al. 2000; Justice et al. 1999) have or are generating a significant number of new genetically defined mice that are actively being archived in existing mouse repositories. In addition, the adoption of gene-editing technologies using TALENs (zinc-finger nucleases, transcription activator-like effector nucleases) and CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats/CRISPR associated system) (Aida et al. 2015; Singh et al. 2015; Sung et al. 2012) by the IMPC and the larger mouse genomics community will contribute to additional expansions to repository inventories.
Informatics infrastructure
An overview of the IMSR system is shown in Fig. 1. Data providers deposit files in a defined format on a private FTP site. The format is a simple tab delimited text file whose fields are specified on an IMSR help page (http://www.findmice.org/participate). An automated process checks the deposit area hourly. New submissions are scanned for formatting and content errors (e.g., missing values or IDs that do not designate valid genes). Any errors are communicated to the providers via email along with instructions for needed fixes and contact information for obtaining assistance. Submissions that pass the error checking process are archived, then processed to improve content against MGD data, indexed by Lucene (http://lucene.apache.org), and made available via our Solr instance (http://lucene.apache.org/solr/). IMSR is a noSQL system; all queries and page generation are supported by Solr/Lucene, which provides very fast response times. IMSR currently uses Solr 5.2 running in a WildFly 8.2 container. A parallel system is used for development and testing.
Overview of IMSR process. (1) Individual repository databases initiate data contribution to IMSR by providing a format-specified text file to IMSR via a private FTP site. Automated checks confirm that the incoming data file is properly formatted and that fields contain the data of correct type. If not, (2a) data are returned with comment to inform the repository of error corrections required for submission. If data pass this automated check, (2b) data are processed to compare them to MGD data (3) standardizing nomenclature to allow better web links and producing nomenclature and new data reports for curator action. Repository data are replaced with a new version (4a) for display on the IMSR website and (4b) curators correct and/or update repository submitted nomenclature and add new data to MGD. The curation results also are returned to the repository (5) for updating their files with the correct nomenclature for the alleles, genes, and strains that they hold in their resource
Concurrently, after a newly submitted data file passes automated checks, comparison with MGD data will reveal any data inconsistencies (e.g., incorrect strain names, mismatched IDs, or gene or allele nomenclature in need of updates). These will be curated and corrections returned to the repository provider so they can update their records. In this manner, repositories become more nomenclature accurate/current and iteratively improve their data; and users benefit from future loads of corrected data being more readily searchable using standard allele, gene, and strain designations.
User interface: the IMSR website
Users access IMSR primarily via a web-based interface (www.findmice.org). Searches can be performed using one or many parameters, including strain parameters, genetic parameters, and repository name/location. Strain parameters include the strain/stock designation, the strain ID, the state in which the strain/resource is maintained (live, cryopreserved embryo, cryopreserved ovary, cryopreserved or freeze-dried sperm, or ES cell line), and the strain type. Genetic parameters include the symbol or name of the phenotypic allele or gene of interest carried in the strain, the relevant allele or gene accession ID, and the type (origin) of the mutation and its chromosomal location. Repository parameters include the name of one or more specific repositories, or the selection of all repositories in a geographical regional location (Fig. 2). The results of a search are returned in tabular format, with each row in the table representing one unique genetic strain from a given repository. [Note, therefore, that if a repository holds a strain in multiple states, the strain is only listed once; but each “state” status is provided]. Search results can be exported in text or Excel format. Figure 3 shows 10 rows of the 29 rows returned when searching for strains carrying mutations in the Stat3 gene (as of May 15, 2015).
IMSR homepage/search page. (www.findmice.org). This web page introduces IMSR and is the search form for IMSR content. The search section has (1) a Quick Search option (green bar located mid-page), where one may type in a single field a mutant allele, gene, or strain and click the search button; (2) a strain search section where one may further specify the “state” (e.g., embryo, live) in which the resource is maintained and the “strain type” which defines how the strain is bred; (3) a Repository search limiting results to one or more specific repositories of choice, or using the map, limit one’s search to a particular continent; and (4) mutation type, specifying how the mutant allele of interest was created (e.g., gene trap, transgenic, targeted)
Example IMSR search result. This screenshot shows the first 10 (of 29) lines returned when searching IMSR for strains with mutations in the Stat3 (signal transducer and activator of transcription 3) gene. Columns shown include (1) “N” for nomenclature, where + is correct; − is incorrect, and ? is unreviewed; (2) Strain Name, each strain name links to the strain description page on the respective repository’s website; (3) Synonyms, other names that this strain has been called, including former names and lab-jargon names; (4) States are how the mouse resource exists—live, cryopreserved embryo, sperm, or ova, or ES cell line; (5) the Repository column provides two links, when available—an email link to user assistance at the repository site and a link to “order” the mouse resource from the repository site; (6) Mutation type, as provided by repository data; (7) Alleles, symbol and name, as provided by repository or enhanced by MGD linking; (8) Genes indicating those mutated in the mouse stock in question; and (7) Strain Type defined by breeding scheme, if known. Buttons to export the current results are found above the strain name column
But how does an investigator who does not know what strain or what mutant he/she may need approach the IMSR? The key is found in the reciprocal links and complementary information contained in MGD (phenotype and disease model data) and the IMSR (strain listings). Figure 4 illustrates the interplay between these sites conceptually. A user directs questions that are phenotype or disease model oriented in nature to the MGD database where he/she can then view the specifics of a mutant phenotype or learn what human disease(s) this mutant is used to model. Each such page in MGD detailing the phenotype and disease models for a given mutant links directly to IMSR for users to physically locate strains or ES cell line resources containing the mutant in question. Similarly, a user of IMSR, when viewing a set of strains and ES cell lines that carry mutations in a particular gene can, for any of those strains, link directly to the MGD detail page describing the phenotype and disease models.
Interplay between MGD and IMSR that maximizes user access. A user searching for a particular strain or mutations in a particular gene can quickly find these resources using the IMSR interface (see Figs. 2, 3 and text). However, if a user does not know the strain or mutant desired, but wants to search for mouse resources on the basis of phenotype or disease model, he/she would start with a phenotype/disease search of MGD. Using either entry point, the user can easily follow links between them. For example, a user who searched MGD for a phenotype of absent cochlear hair cells or the human disease Pendred Syndrome would view phenotype detail pages such as the one shown for Slc26a4 pdsm (top panel). From this page, a link to IMSR (red circle) will lead the user to IMSR for a list of strains and cell lines available with Slc26a4 mutations (bottom panel). The link shown from IMSR (red circle) will lead to the relevant detailed phenotype/disease page in MGD
Data standards and challenges in maintaining and updating IMSR
The largest challenge of maintaining IMSR is the variability in data quality and completeness among the data files submitted to IMSR from different repositories. Although there is documentation about data fields and required format, data received, particularly from smaller repositories with less sophisticated informatics infrastructure, may be incomplete, or the repositories may not have some critical information from the original source that generated the mice. However, for any given strain holding submitted in the repository file, if the minimum data fields are provided (strain ID, strain name, state, strain type), that strain can display in the IMSR website, but only limited links to other information resources will be possible.
A second challenge for IMSR is the use of non-standard nomenclature in the gene, mutant allele, and strain designations provided to IMSR. In processing incoming files, IMSR scripts are run that attempt some automatic data field completion (e.g., if the repository provided a nomenclature-correct mutant allele, but left the gene field blank, the correct gene can be inferred). Other scripts compare incoming data with MGD data to allow withdrawn nomenclature or nomenclature synonyms to be replaced with the correct symbols and names on the IMSR website. These automatic corrections allow links to gene and allele data that would otherwise not be possible.
Many nomenclature errors cannot be easily interpreted as described above. These are displayed on the IMSR website ‘as is.’ A curator reviews logs of data errors from incoming repository files and returns corrections to the repository, where possible. It remains incumbent upon the repository to update their own holdings’ database with corrected nomenclature and IDs. This feedback is intended to improve the repository’s own site, as well as to ensure that the next data file provided to IMSR is correct and does not again appear in the error log.
Perspective on mouse repositories
Mouse resources are important for all who use mouse as an experimental system. The ability to obtain genetically defined mouse resources of known health status is key for producing results that are reproducible and can be built upon in future work. Mouse repositories focus their efforts on providing the highest quality, genetically tested, and strain background-defined resources possible. The IMSR facilitates their work by assisting investigators worldwide with access to those resources, wherever they exist. The last decade has seen an explosion of new mouse resources produced aided by new genetic technologies. These are making their way into repositories, with a rapid uptake by experimental scientists. The work of the global repository network and the IMSR remain important pieces of the infrastructure fabric for biological and disease model research of the future.
References
Aida T, Chiyo K, Usami T, Ishikubo H, Imahashi R, Wada Y, Tanaka KF, Sakuma T, Yamamoto T, Tanaka K (2015) Cloning-free CRISPR/Cas system facilitates functional cassette knock-in in mice. Genome Biol 16:87
Arnold CN, Barnes MJ, Berger M, Blasius AL, Brandl K, Croker B, Crozat K, Du X, Eidenschenk C, Georgel P, Hoebe K, Huang H, Jiang Z, Krebs P, La Vine D, Li X, Lyon S, Moresco EM, Murray AR, Popkin DL, Rutschmann S, Siggs OM, Smart NG, Sun L, Tabeta K, Webster V, Tomisato W, Won S, Xia Y, Xiao N, Beutler B (2012) ENU-induced phenovariance in mice: inferences from 587 mutations. BMC Res Notes 5:577
Brown SD, Moore MW (2012) The international mouse phenotyping consortium: past and future perspectives on mouse phenotyping. Mamm Genome 23:632–640
Eppig JT, Strivens M (1999) Finding a mouse: The International Mouse Strain Resource (IMSR). Trends Genet 15:21–81
Eppig JT, Bult CJ, Kadin JA, Richardson JE, Blake JA, Anagnostopoulos A, Baldarelli RM, Baya M, Beal JS, Bello SM, Boddy WJ, Bradt DW, Burkart DL, Butler NE, Campbell J, Cassell MA, Corbani LE, Cousins SL, Dahmen DJ, Dene H, Diehl AD, Drabkin HJ, Frazer KS, Frost P, Glass LH, Goldsmith CW, Grant PL, Lennon-Pierce M, Lewis J, Lu I, Maltais LJ, McAndrews-Hill M, McClellan L, Miers DB, Miller LA, Ni L, Ormsby JE, Qi D, Reddy TB, Reed DJ, Richards-Smith B, Shaw DR, Sinclair R, Smith CL, Szauter P, Walker MB, Walton DO, Washburn LL, Witham IT, Zhu Y, Mouse Genome Database Group (2005) The mouse genome database (MGD): from genes to mice—a community resource for mouse biology. Nucleic Acids Res 33:D471–D475
Goldowitz D, Frankel WN, Takahashi JS, Holtz-Vitaterna M, Bult C, Kibbe WA, Snoddy J, Li Y, Pretel S, Yates J, Douglas J, Swanson DJ (2004) Large-scale mutagenesis of the mouse to understand the genetic bases of nervous system structure and function. Brain Res Mol Brain Res 132:105–115
Hrabé de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, Alessandrini F, Jakob T, Fuchs E, Kolb H, Kremmer E, Schaeble K, Rollinski B, Roscher A, Peters C, Meitinger T, Strom T, Steckler T, Holsboer F, Klopstock T, Gekeler F, Schindewolf C, Jung T, Avraham K, Behrendt H, Ring J, Zimmer A, Schughart K, Pfeffer K, Wolf E, Balling R (2000) Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet 25:444–447
Justice MJ, Noveroske JK, Weber JS, Zheng B, Bradley A (1999) Mouse ENU mutagenesis. Hum Mol Genet 8:1955–1963
Li Y, Klena NT, Gabriel GC, Liu X, Kim AJ, Lemke K, Chen Y, Chatterjee B, Devine W, Damerla RR, Chang C, Yagi H, San Agustin JT, Thahir M, Anderton S, Lawhead C, Vescovi A, Pratt H, Morgan J, Haynes L, Smith CL, Eppig JT, Reinholdt L, Francis R, Leatherbury L, Ganapathiraju MK, Tobita K, Pazour GJ, Lo CW (2015) Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature 521:520–524
Mallon AM, Iyer V, Melvin D, Morgan H, Parkinson H, Brown SD, Flicek P, Skarnes WC (2012) Accessing data from the international mouse phenotyping consortium: state of the art and future plans. Mamm Genome 23:641–652
Nolan PM, Peters J, Vizor L, Strivens M, Washbourne R, Hough T, Wells C, Glenister P, Thornton C, Martin J, Fisher E, Rogers D, Hagan J, Reavill C, Gray I, Wood J, Spurr N, Browne M, Rastan S, Hunter J, Brown SD (2000) Implementation of a large-scale ENU mutagenesis program: towards increasing the mouse mutant resource. Mamm Genome 11:500–506
Singh P, Schimenti JC, Bolcun-Filas E (2015) A mouse geneticist’s practical guide to CRISPR applications. Genetics 199:1–15
Sung YH, Baek I-J, Seong JK, Kim J-S, Lee H-W (2012) Mouse genetics: catalogue and scissors. BMB Rep 45:686–692
Wilkinson P, Sengerova J, Matteoni R, Chen CK, Soulat G, Ureta-Vidal A, Fessele S, Hagn M, Massimi M, Pickford K, Butler RH, Marschall S, Mallon AM, Pickard A, Raspa M, Scavizzi F, Fray M, Larrigaldie V, Leyritz J, Birney E, Tocchini-Valentini GP, Brown S, Herault Y, Montoliu L, de Angelis MH, Smedley D (2010) EMMA–mouse mutant resources for the international scientific community. Nucleic Acids Res 38:D570–D576
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Eppig, J.T., Motenko, H., Richardson, J.E. et al. The International Mouse Strain Resource (IMSR): cataloging worldwide mouse and ES cell line resources. Mamm Genome 26, 448–455 (2015). https://doi.org/10.1007/s00335-015-9600-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-015-9600-0