Abstract
Two large-scale phenotyping efforts, the European Mouse Disease Clinic (EUMODIC) and the Wellcome Trust Sanger Institute Mouse Genetics Project (SANGER-MGP), started during the late 2000s with the aim to deliver a comprehensive assessment of phenotypes or to screen for robust indicators of diseases in mouse mutants. They both took advantage of available mouse mutant lines but predominantly of the embryonic stem (ES) cells resources derived from the European Conditional Mouse Mutagenesis programme (EUCOMM) and the Knockout Mouse Project (KOMP) to produce and study 799 mouse models that were systematically analysed with a comprehensive set of physiological and behavioural paradigms. They captured more than 400 variables and an additional panel of metadata describing the conditions of the tests. All the data are now available through EuroPhenome database (www.europhenome.org) and the WTSI mouse portal (http://www.sanger.ac.uk/mouseportal/), and the corresponding mouse lines are available through the European Mouse Mutant Archive (EMMA), the International Knockout Mouse Consortium (IKMC), or the Knockout Mouse Project (KOMP) Repository. Overall conclusions from both studies converged, with at least one phenotype scored in at least 80 % of the mutant lines. In addition, 57 % of the lines were viable, 13 % subviable, 30 % embryonic lethal, and 7 % displayed fertility impairments. These efforts provide an important underpinning for a future global programme that will undertake the complete functional annotation of the mammalian genome in the mouse model.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Sequencing a genome will soon be routinely achieved in less than a week. Nevertheless, despite advances in our ability to decipher and annotate genomes, our understanding of the functions of genes is limited. With its highly developed genetics, the mouse has become the mammalian model of choice for the study of gene function (Collins et al. 2007). Almost one third of the known mouse genes representing predictive forms of a coding sequence are still not well annotated for functions. A third of mouse genes possess some functional annotation, reflecting the analysis of point mutations, though not always related to loss-of-function effects. The analysis of the remaining third has depended on the specific interests and expertise of investigators and not, in most cases, on a comprehensive phenotyping approach.
Several initiatives have combined effort over the last few years to attempt a systematic and comprehensive functional assessment of all mouse genes. Based on a roadmap discussed in 2004 (Austin et al. 2004; Auwerx et al. 2004), a series of mouse programmes have been set up with the goal of generating a compendium of mutations in embryonic stem (ES) cells, to produce the mutant mouse lines, to phenotype them, and to make these new resources available to the scientific community (Table 1). The first element of this vision, the construction of a mutant ES cell resource for every mouse gene, has led to the development of the International Knock-out Mouse Consortium (IKMC), which brings together the various programmes funded in Europe (European Conditional Mouse Mutagenesis, EUCOMM), the US (Knock-Out Mouse Project, KOMP, and Texas A&M Institute for Genomic Medicine, TIGM), Canada (North American Conditional Mouse Mutagenesis Project, NorCOMM), and Asia (The Asian Mouse Mutagenesis and Resource Association, AMMRA). This consortium began its work in 2006 and aims to generate a complete resource of reporter-targeted alleles in C57BL/6 N (B6 N) mouse ES cells. The C57BL/6 genetic background is considered to be the ideal background for large-scale phenotyping with a highly characterized genome and one of the best characterised inbred strains, although resistant to certain complex traits such as tumour formation. The design and construction of the conditional targeting constructs was performed through a well-defined pipeline (Skarnes et al. 2011).
The European Mouse Clinics organised the EUMORPHIA programme in order to develop new mouse phenotyping approaches from 2002 to 2006. This led to the definition of the European Mouse Phenotyping Resource of Standardised Screens (EMPRESS), with standard operating procedures (SOPs) for mouse functional analysis (Brown et al. 2005). Using the knowledge gained and the acquired experience of large-scale chemical mutagenesis programmes (Hrabé de Angelis et al. 2000; Nolan et al. 2000), the European Mouse Disease Clinic (EUMODIC) brought together four European mouse clinics, the MRC in Harwell (UK), the German Mouse Clinic in Munich (Gailus-Durner et al. 2005), the Institut Clinique de la Souris (ICS) in Strasbourg-Illkirch (France), and the Wellcome Trust Sanger Institute in Hinxton, which also launched the Mouse Genetics Project (SANGER-MGP) to produce large-scale standardized mouse phenotyping data. With the EUCOMM programme commencing a year before to produce mutant ES cells in up to 8,000 mouse genes, it was a perfect opportunity to take advantage of the targeted ES cells generated by this consortium to generate mouse mutant lines and to phenotype them.
After four years of activity both programmes together produced 665 new mouse lines and the number continues to increase. The high-throughput production and phenotyping assist in deciphering the function of genes with no prior functional annotation and enhance the understanding of genes with already known functions, moving a step further in building an encyclopaedia of mammalian gene function in the mouse.
From mutant ES cells to mouse lines
In order to carry out standardized and comprehensive phenotyping, both the EUMODIC and the SANGER-MGP programmes preferentially used ES cells from EUCOMM and KOMP programmes. These programmes started in 2006 to produce mutant ES cells with a worldwide target of 16,500 mouse genes. They used standardized procedures to generate mutant loci for genes of interest in B6 N ES cells. This created the perfect resource for the production of isogenic mutant lines in a standardized way and to carry out phenotyping.
As a consequence, 83 % (665/799) of the mouse lines were generated for the SANGER-MGP and the EUMODIC programmes from the EUCOMM and KOMP ES cell resources, respectively. The majority of mouse lines were carrying the knockout first allele named “tm1a” in which the transcription of a locus of interest is blocked by the insertion of a lacZ minigene with a strong polyA sequence just before a critical exon. A neomycin selection cassette was found after the reporter sequence. Its expression was driven either by the endogenous gene promoter (promoterless tm1a) with the β-galactosidase fused to the neomycin resistance protein with a T2A cleavage site, or by a specific promoter (promoter-driven tm1a) (Friedel et al. 2007; Skarnes et al. 2011). The tm1a allele creates a premature termination of the transcript with an interruption of the coding sequence and in most cases is associated with mRNA decay and the expression of the reporter gene. The knockout first conditional ready allele is certainly very valuable as it is possible to convert the tm1a allele to a conditional ready tm1c allele and phenotype a lethal embryonic gene by breeding the homozygous conditional mouse with a cell- or tissue-specific Cre or inducible CreERT2 line (Birling et al. 2009). Another advantage is that the lacZ sequence from the tm1a alleles is driven by the endogenous promoter allowing evaluation of cells that express the targeted gene. Furthermore, the ES cell resources were based on B6 N ES cells; this allows the generation of mouse lines in a pure genetic background as the chimeras were bred to that strain. More detailed information regarding EUCOMM and KOMP clones but also TIGM and NorCOMM mutant ES cell lines is found at the IKMC web portal (http://www.knockoutmouse.org/about) and in detail elsewhere (Ringwald et al. 2011).
The targeted genes from the ES cell resources were chosen with input from the scientific community. They were also selected because of absence of an available (conditional) mouse line in the public domain or phenotyping data. Mutant ES cells were produced, validated, and verified prior to injection at the mouse clinics. Both phenotyping programmes were keen on evaluating and improving the applicability of the mouse production and phenotyping schemes. The remaining mouse models (134/799) came from individual laboratories. They were chosen for their scientific interest, with the commitment of their owner to make the phenotyping data available as it became available. They were also pilot experiments for the phenotyping pipelines. Interestingly, different types of alleles were analysed, including null alleles, gene trap, ENU-induced mutations, large-scale deletion, and transchromosomic mice, underlying the general interest in the broad-based phenotyping effort.
The mutant mouse lines were generated from the ES cell resources using different injection strategies, using either BALB/cN or C57BL/6Brd-Tyr c-Brd blastocyst donors, with the aim of generating high rates of germline transmission (GLT). The injection parameters (e.g., number of injected cells per embryo, culture media conditions) were optimised. A minimum of 35 % and up to 70 % of GLT was obtained in the different centres. In order to simplify the breeding schemes using C57BL/6 N blastocysts, it was decided that the dominant agouti coat colour gene would be restored in the basic JM8 cells by targeted repair of the B6 nonagouti mutation (Pettitt et al. 2009). The EUCOMM and KOMP ES cell resource contains different sublines of the original ES cell line JM8 (JM8.F4, JM8.N6), with additional lines carrying the restored agouti allele (JM8A3.N1 and JM8A1.N3). Pettitt et al. (2009) presented a figure illustrating the coat colour of mice and their offspring when creating mouse lines using the different JM8 sublines. Eighty-nine percent of the mouse lines generated from the IKMC consortium ES cells and phenotyped in the EUMODIC consortium originated from the nonagouti B6 N ES cells. In total, 665 mouse lines were generated from the mutant ES cell resources. At the ICS, germline transmission was achieved, with a median time for the age of the mouse chimera of 109 days and a minimum of 65 days; 95.5 % of the GLTs were obtained by 166 days.
The tm1a allele or knockout first allele is potentially a nonexpressed (null) form in which transcription of the targeted gene is stopped. However, RT-qPCR performed on different targeted mouse models showed that in some mouse models or ES cells the polyA did not entirely stop the transcription (data not shown). In a few cases a splicing of the selection cassette was observed resulting in production of a low-level wild-type transcript (data not shown). The potential of generating a knockdown instead of a real knockout exists, even if the incidence of detecting a small amount of wild-type protein is not truly assessed. As a result of this, the IMPC consortium has decided to remove the floxed coding region prior to phenotypic analysis of future projects (especially for promoter-containing targeting cassettes as the promoter-driven neomycin is also removed) to be sure that true knockout models are being phenotyped.
From germline transmission to cohorts
Generating the phenotyping cohort is a critical step in the process of phenotyping, from the scientific and welfare perspectives. Space and costs are bottleneck issues during the process and are affected by several components, including cohort size, age-matching animals, control strategy, genetic background, power of the tests, and statistical management of the data.
The genetic background strongly contributes to phenotypic variations with a major impact on large biological functions in genetically engineered mice. It could affect the expression or penetrance of a given phenotype (Champy et al. 2008; Doetschman 2009; Fitch et al. 2003; Ramírez-Solis et al. 1993; Threadgill et al. 1995). To minimize this impact, both high-throughput phenotyping projects established the mutations on a pure B6 N background. The direct comparison of mutants with an appropriately matched control population of mice therefore was possible. Not only was the background uniform, but as a consequence of the breeding scheme, the controls and the mutants differed only by a limited number of breeding generations (Fig. 1). These strategies offered two advantages: first, the production of a baseline data collection to evaluate the variance, and, second, the facilitation of the interlaboratory comparison. However, one pitfall of the uniform genetic background is the potential limit on the consequences of a given mutation. Beside the ES cell resource, both programmes analysed a limited number of mouse lines coming from different laboratories with a mixed or other background strain, and in that particular case, littermates or appropriate inbred mice were used as controls.
Flowchart for ES cells injection and mouse breeding strategies for cohort production. Targeted ES cells were injected to generate chimeric animals. After germline transmission of the mutant allele, the heterozygous mice were bred for pedigree expansion, archiving, distribution, and cohort production. Depending on the fertility score, heterozygous or homozygous matings were used to generate cohorts of animals for mouse phenotyping
The generation of cohorts on a large scale raises considerable logistical issues and has proven to be the major rate-limiting factor for high-throughput phenotyping. The cohort size is driven largely by the nature of the tests, and, in order to detect a significant effect (if any exists), some tests required a consistent age-matched cohort. Within EUMODIC and MGP programmes, from seven to ten mutant mice per gender had been fixed to assess the variance in phenotypes according to the power of the phenotyping tests. To produce such cohorts, the size of the breeding colonies was based on (1) this age-matched cohort requirement, (2) the reproductive strain characteristics of the parental strain used to generate the mutant strain, and (3) the various breeding schemes used between phenotyping centres.
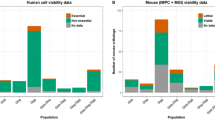
All the mouse production and procedures were performed according to local ethical committee guidelines. Determination of animal welfare is intrinsically linked to phenotype outcome; therefore, animal welfare assessments were performed through centre-specific procedures. Nevertheless, the major bottleneck was found to be expansion of the colony, and several mouse mutant lines displayed impaired viability and fertility. Thus, embryonic or perinatal lethality and fertility were assessed as part of the overall phenotypic analysis during mouse production. Data were recorded first for the viability of homozygous offspring at weaning stage (2–4 weeks old) from heterozygous intercrosses. Viability assessment was done by genotyping a minimum of 28 offspring in order to be 95 % confident that the probability of homozygous survival is inferior or equal to 40 %. From these criteria, three outcomes were possible per mutant strain: (i) embryonic or perinatal lethality if no homozygous progeny was detected from the 28 littermates, (ii) subviability if the number of homozygotes detected was between 0 and 13 % of the 28 littermates from heterozygote intercrosses, and (iii) viability if the number of homozygotes was above 13 %. Unexpectedly, only 57 % of the 461 mouse mutant lines analysed was viable based on the results of the two programmes, with 13 % subviable and 30 % showing homozygous lethality (Table 2). The homozygous lethality was strongly dependent on the tm1a subtype, with “promoterless” targeted loci displaying 60 % of lethality compared to 28 % observed in “promoter-driven” mutant loci (χ2 test P = 2.5 × 10−11). In the “promoterless” targeted loci, the gene should be expressed in ES cells to drive the neomycin resistance. This correlation presumably reflects that loci expressed in ES cells have a greater probability to control critical embryonic processes and to induce lethality. This percentage of lethal genes is comparable to that observed in different chemical mutagenesis screens (Boles et al. 2009; Ching et al. 2010; Hentges et al. 2006; Kile et al. 2003; Stottmann and Beier 2010; Stottmann et al. 2011). The lethal lines and some subviable lines were further phenotyped as heterozygotes.
Fertility of homozygous genetically altered mouse lines was assessed by mating at least two sexually mature mice from both genders. A mutant line was recorded as fertile using homozygous intercrosses, when offspring are generated regardless of the viability of the offspring. If fertility is uncertain in homozygous intercrosses, homozygous mutant mice are mated to nonhomozygous mice (either heterozygotes or wild types; Fig. 1). From this basic fertility assessment, mutant lines are recorded as (1) “generates offspring” if pups are produced or (2) “abnormal fertility” if no living pups are generated. Fertility assessment was also done on heterozygotes for lethal lines and labelled male, female, or both. The third outcome, not applicable (N/A), is recorded for particular classes such as imprinted genes and homozygous lethal lines. Current data indicate that overall 7 % of the mutant lines (23 of 330 tested) showed abnormal fertility.
The production of full-size cohorts was thus often the result of a compromise by finding particular homozygous mutants with impaired fertility or viability and also by centre-specific logistic constraints. As a consequence, partial-sized cohorts (on average, 3 cohorts per line) were often passed through each pipeline to reach the minimum number per gender (7 mice). Figure 1 summarizes the workflow used by the centres to optimize the mouse production for the phenotyping pipeline, which balanced the cohort size production requirement, the viability and fertility phenotype annotations, and mouse distribution. All the mutant models were archived and made available for the scientific community through the European Mouse Mutant Archive (EMMA) repository (Wilkinson et al. 2010) and KOMP repositories or directly from the SANGER-MGP.
From mouse to phenotypes
The aim of the two phenotyping programmes was to perform a comprehensive phenotyping workflow to generate data covering most body systems, physiology, and behaviour. The EUMODIC and SANGER-MGP programmes were set up independently to carry out a comprehensive analysis and a screen of mouse mutant lines, respectively.
The EUMODIC programme used the SOPs developed within the EUMORPHIA consortium (www.eumorphia.org; Brown et al. 2005). It was shaped around two independent pipelines: the first one was devoted to morphology, metabolism, and skeletal and cardiovascular systems, while the second was oriented toward neurobehavioural and sensory systems, haematology, biochemistry, and baseline immune responses (Eumodic consortium, unpublished). The analysis began at age 9 weeks and was completed by 15 weeks of age. Its design was based on the use of two separate cohorts of mice, each composed of at least seven mutants of each gender, to detect differences in physiology or diseases, recognising that gender may have a considerable impact upon disease prevalence. It was also recommended that control mice be analysed through the phenotyping pipelines at the same time as mutants. Usually C57BL/6 N mice have been used. Mice should be born within a timeframe of 7–10 days. The phenotyping assays that have been chosen for the EMPReSSslim [European Mouse Phenotyping Resource of Standardised Screens (EMPReSS) Slim] workflow are limited, but robust, providing a relatively broad-based first pass phenotype assessment, both high-throughput and cost-effective. All SOPs are available on the EuroPhenome web site and are based on the EUMORPHIA programme (Brown et al. 2005, 2009; Morgan et al. 2010). In total, two cohorts of at least seven males and seven females were analysed through the 20 platforms of pipelines 1 and 2 (Table 3), and 413 phenotyping variables and 146 environmental or experimental metadata were recorded (Table 4).
The SANGER-MGP pipeline was designed as a single pipeline starting at 5 weeks of age with a high-fat diet challenge, and followed by a battery of tests from week 4 to 16, as shown in Fig. 2 and Table 3. The pipeline was designed to identify a variety of models from obesity to dysmorphology with a series of 13 wide-ranging platform tests, and record a number of variables for phenotypic assessment as well as additional metadata impacting conditions or equipment. Groups of mutant animals (up to 7 males and 7 females per line) went through as they were produced, along with one control cohort each week for each genetic background. The pipeline was further enriched by additional procedures devoted to specific functions such as a challenge for immune or auditory brainstem response measurements that could be analysed during the pipeline or with a new batch of mutant cohorts (Table 5).
Schematic overview of the EUMODIC and MGP pipelines. The Sanger MGP pipeline started at the age of 4 weeks and ended at 16 weeks of age, whereas the EUMODIC programme encompasses two pipelines, 1 and 2, which started at 9 weeks and were completed after 7 and 6 weeks, respectively. The phenotyping platforms used in these pipelines are indicated
The two phenotyping programmes focused on basic physiological aspects, with platforms operating with nearly identical or similar procedures for 11 and slightly different ones for 8 (Table 3) with a set of common variables and metadata. Statistical analyses have been set up based on the way cohorts and controls were produced to ensure the use of appropriate tests to detect the phenodeviants when they exist. Two main approaches were applied: (1) classical inferential statistics based on the use of large baseline control sets, and (2) for the partial phenotyping group, the reference range approach was preferred, using the baseline data collection which ensured a simple and robust strategy. The reference range approach was preferentially used by the SANGER-MGP approach irrespective of the cohort size. The analysis of data also helped to refine the tests and to reduce the number of animals used in the cohorts (Karp et al. 2010).
Even though the precise timing and the local organization of the workflow varied, and differences between platforms exist between the two phenotyping programmes and the four centres involved, the efficiency of the platforms in detecting phenotype is quite similar (Eumodic consortium, unpublished). Both phenotyping pipelines were successful in identifying phenotypes in mutants for genes for which there was phenotyping annotation for at least one allele already available. Sixteen of 30 mutants analysed in the EUMODIC programme displayed phenotyping overlaps to annotations found in the Mouse Genome Informatics database (http://www.informatics.jax.org/) (Eumodic consortium, unpublished).
Taking into account the fertility and viability annotations, both programmes revealed at least one phenotype in about 83.8 % (EuroPhenome) and 76.5 % (SANGER-MGP) of the mutant lines, demonstrating the power of comprehensive phenotyping in defining functions for any gene of interest. A few additional platform screens were stopped or abandoned in the SANGER-MGP programme and replaced with new tests (Table 5). With this new panel of tests, the SANGER-MGP programme increased the coverage in phenotyping particular aspects not detailed within the common shared platforms. Where homozygote mutants were not viable, both programmes have analysed heterozygotes. Accordingly, the EUMODIC programme had a phenotypic hit rate of 71.7 % in this class, underlining the potential for the annotation of genes through heterozygote analysis.
Future perspectives
In conclusion, the main objective of the two efforts was achieved with the generation of nearly 800 mouse models, which are available and accompanied by a broad-based phenotype analysis. Furthermore, both programmes reach similar conclusions on mutant lethality, finding that around 30 % of mouse mutants are not viable. Both programmes highlighted the efficiency of the broad-based phenotyping approaches, identifying at least one phenotype in around 80 % of the mouse mutants. The mouse clinics have provided a new set of data on gene function to the scientific community while improving the throughput and reducing the cost of the analysis. The results summarized here provide many logistical, operational, and scientific lessons that will be vital as we begin the next step in undertaking a global project of exploration of the function of genes and generation of an encyclopaedia of the mammalian genome by the IMPC.
Nevertheless, additional resources are required, most importantly the completion of the project of generating mutant ES cells for all loci in the mouse genomes: not all the genes are available yet in the IKMC. New initiatives have already begun with three new programmes—The EUCOMM-Tools for Functional Annotation of the Mouse Genome (EUCOMMTools), the Knockout Mouse Phenotyping Programme (KOMP2), and Norcommt2ls—to generate a final series of mutant ES cells, along with a new set of genetic tools for conditional genetics. Since 2011, miR knockout (KO) ES cells are also available on the IKMC web site. Prosser et al. (2011) have developed a resource of vectors and ES cells for targeted deletion of microRNAs in mice and show how to generate, using Recombinase Mediated Cassette Exchange (RMCE) from this miR KO allele, a reporter of the miR promoter activity, the conditional KO, and how to mutate one or other miR organised within the miR clusters. Further resources will be needed for long noncoding RNAs, for point mutations, and for copy number variants, and more generally to decipher the regulatory mechanism of the transcriptional and genome organisation. In addition, it will be important to consider routes to explore heterozygous lethal or haploinsufficient mutations that escape current analyses.
In the two programmes discussed, the mutants generated so far and analysed were almost all derived from one allele, the tm1a allele. A limitation of this approach is the efficient transcription stop due to the presence of the polyA. We know from a series of experiments in the different clinics that some tm1a alleles do not lead to full expected inactivation of the targeted gene. One way to circumvent the problem is to use the tm1b allele carrying the deletion of the critical exon(s) without the removal of the lacZ sequence (or the lacZ and neo sequences in the promoterless mutants). To obtain such a tm1b allele, different universal deleters are available, but some deleters are likely to be preferred. For example, the Gt(ROSA)26Sortm1(ACTB-cre,-EGFP)Ics deleter eliminates one generation of breeding due to the maternal accumulation of the Cre in the oocytes allowing the accumulation of the recombinase during oogenesis and the direct recovery of individuals carrying the deletion without the Cre transgene (Birling et al. 2011). In addition, the line was developed on a pure C57BL/6 N genetic background and the presence of the transgene can be genotyped by GFP fluorescence.
The main outcomes of the first set of mutants that were analysed are the high level of homozygote lethality observed with over 30 % of the lines studied and the high level of phenotype annotations associated with heterozygous mutants. Nevertheless, a number of additional approaches should be considered. First, the lacZ pattern of the mutant allele should be explored. The use of tm1a or tm1b allele’s lacZ reporter enables us to study the expression of the gene with good sensitivity in heterozygotes. Of course, an inconvenience of this strategy is that regulatory elements or other genomic elements (e.g., ncRNA, miRNA) in the intron of the gene are modified as well. In addition, the locus might code for various isoforms with different promoters. If the isoform expressed in the organ of interest is different from the isoform targeted and if it uses a different (endogenous) promoter, the lacZ will not be expressed as expected. Second, the phenotyping pipeline should also integrate an embryonic screen to determine key steps of embryonic development. Some centres have already developed an embryonic lethal screening pipeline on a limited number of lines with some success.
The EUMODIC and SANGER-MGP programmes pioneered the high-throughput production and comprehensive analysis of mice knockouts. Considerable experience has been gained in the logistics and operation of phenotyping pipelines that will underpin future developments in international phenotyping programmes. For example, much information has been gathered on the effectiveness and sensitivity of phenotyping platforms that will prove valuable in the design of future pipelines, including the introduction of new assays to cover physiological systems. Along with the new partners of the IMPC, a new adult pipeline has been developed and the establishment of a unique and high-throughput embryonic pipeline is under discussion. As part of this process, additional assays for immune systems, inflammation, respiration, cognition, and behaviour have all been considered. In addition, building the research infrastructure capacities required in Europe for these large-scale efforts, as well as for access of individual researchers to high-throughput phenotyping, is the task of the pan-European Infrafrontier programme, which has been prioritized by ESFRI (European Strategy Forum on Research Infrastructures). Infrafrontier also secures sustainable access to mouse models archived in EMMA. Similar efforts are underway in North America and in Asia. With these new developments, we can expect that the upcoming large-scale phenotyping pipeline operated by the IMPC will prove even more effective in delivering broad-based phenotype information on a wide variety of body systems and physiological functions.
References
Austin C, Battey J, Bradley A, Bucan M, Capecchi M, Collins F, Dove W, Duyk G, Dymecki S, Eppig J, Grieder F, Heintz N, Hicks G, Insel T, Joyner A, Koller B, Lloyd K, Magnuson T, Moore M, Nagy A, Pollock J, Roses A, Sands A, Seed B, Skarnes W, Snoddy J, Soriano P, Stewart D, Stewart F, Stillman B, Varmus H, Varticovski L, Verma I, Vogt T, von Melchner H, Witkowski J, Woychik R, Wurst W, Yancopoulos G, Young S, Zambrowicz B (2004) The knockout mouse project. Nat Genet 36:921–924
Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, Chambon P, Cox R, Davidson D, Davies K, Duboule D, Forejt J, Granucci F, Hastie N, de Angelis M, Jackson I, Kioussis D, Kollias G, Lathrop M, Lendahl U, Malumbres M, von Melchner H, Müller W, Partanen J, Ricciardi-Castagnoli P, Rigby P, Rosen B, Rosenthal N, Skarnes B, Stewart A, Thornton J, Tocchini-Valentini G, Wagner E, Wahli W, Wurst W (2004) The European dimension for the mouse genome mutagenesis program. Nat Genet 36:925–927
Birling M, Gofflot F, Warot X (2009) Site-specific recombinases for manipulation of the mouse genome. Methods Mol Biol 561:245–263
Birling MC, Dierich A, Jacquot S, Hérault Y, Pavlovic G (2011) Highly-efficient, fluorescent, locus directed Cre and flpo deleter mice on a pure C57BL/6 N genetic background. Genesis 50:482–489
Boles MK, Wilkinson BM, Maxwell A, Lai L, Mills AA, Nishijima I, Salinger AP, Moskowitz I, Hirschi KK, Liu B, Bradley A, Justice MJ (2009) A mouse chromosome 4 balancer ENU-mutagenesis screen isolates eleven lethal lines. BMC Genet 10:12
Brown S, Chambon P, de Angelis M (2005) EMPReSS: standardized phenotype screens for functional annotation of the mouse genome. Nat Genet 37:1155
Brown SD, Wurst W, Kühn R, Hancock JM (2009) The functional annotation of mammalian genomes: the challenge of phenotyping. Annu Rev Genet 43:305–333
Champy MF, Selloum M, Zeitler V, Caradec C, Jung B, Rousseau S, Pouilly L, Sorg T, Auwerx J (2008) Genetic background determines metabolic phenotypes in the mouse. Mamm Genome 19:318–331
Ching YH, Munroe RJ, Moran JL, Barker AK, Mauceli E, Fennell T, Dipalma F, Lindblad-Toh K, Abcunas LM, Gilmour JF, Harris TP, Kloet SL, Luo Y, McElwee JL, Mu W, Park HK, Rogal DL, Schimenti KJ, Shen L, Shindo M, Shou JY, Stenson EK, Stover PJ, Schimenti JC (2010) High resolution mapping and positional cloning of ENU-induced mutations in the Rw region of mouse chromosome 5. BMC Genet 11:106
Collins FS, Rossant J, Wurst W, Consortium IMK (2007) A mouse for all reasons. Cell 128:9–13
Doetschman T (2009) Influence of genetic background on genetically engineered mouse phenotypes. Methods Mol Biol 530:423–433
Fitch K, McGowan K, van Raamsdonk C, Fuchs H, Lee D, Puech A, Hérault Y, Threadgill D, Hrabé de Angelis M, Barsh G (2003) Genetics of dark skin in mice. Genes Dev 17:214–228
Friedel RH, Seisenberger C, Kaloff C, Wurst W (2007) EUCOMM-the European conditional mouse mutagenesis program. Brief Funct Genomic Proteomic 6:180–185
Gailus-Durner V, Fuchs H, Becker L, Bolle I, Brielmeier M, Calzada-Wack J, Elvert R, Ehrhardt N, Dalke C, Franz TJ, Grundner-Culemann E, Hammelbacher S, Hölter SM, Hölzlwimmer G, Horsch M, Javaheri A, Kalaydjiev SV, Klempt M, Kling E, Kunder S, Lengger C, Lisse T, Mijalski T, Naton B, Pedersen V, Prehn C, Przemeck G, Racz I, Reinhard C, Reitmeir P, Schneider I, Schrewe A, Steinkamp R, Zybill C, Adamski J, Beckers J, Behrendt H, Favor J, Graw J, Heldmaier G, Höfler H, Ivandic B, Katus H, Kirchhof P, Klingenspor M, Klopstock T, Lengeling A, Müller W, Ohl F, Ollert M, Quintanilla-Martinez L, Schmidt J, Schulz H, Wolf E, Wurst W, Zimmer A, Busch DH, de Angelis MH (2005) Introducing the German Mouse Clinic: open access platform for standardized phenotyping. Nat Methods 2:403–404
Hentges KE, Nakamura H, Furuta Y, Yu Y, Thompson DM, O’Brien W, Bradley A, Justice MJ (2006) Novel lethal mouse mutants produced in balancer chromosome screens. Gene Expr Patterns 6:653–665
Hrabé de Angelis MH, Flaswinkel H, Fuchs H, Rathkolb B, Soewarto D, Marschall S, Heffner S, Pargent W, Wuensch K, Jung M, Reis A, Richter T, Alessandrini F, Jakob T, Fuchs E, Kolb H, Kremmer E, Schaeble K, Rollinski B, Roscher A, Peters C, Meitinger T, Strom T, Steckler T, Holsboer F, Klopstock T, Gekeler F, Schindewolf C, Jung T, Avraham K, Behrendt H, Ring J, Zimmer A, Schughart K, Pfeffer K, Wolf E, Balling R (2000) Genome-wide, large-scale production of mutant mice by ENU mutagenesis. Nat Genet 25:444–447
Karp NA, Baker LA, Gerdin AK, Adams NC, Ramírez-Solis R, White JK (2010) Optimising experimental design for high-throughput phenotyping in mice: a case study. Mamm Genome 21:467–476
Kile BT, Hentges KE, Clark AT, Nakamura H, Salinger AP, Liu B, Box N, Stockton DW, Johnson RL, Behringer RR, Bradley A, Justice MJ (2003) Functional genetic analysis of mouse chromosome 11. Nature 425:81–86
Morgan H, Beck T, Blake A, Gates H, Adams N, Debouzy G, Leblanc S, Lengger C, Maier H, Melvin D, Meziane H, Richardson D, Wells S, White J, Wood J, de Angelis MH, Brown SD, Hancock JM, Mallon AM, Consortium E (2010) EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res 38:D577–D585
Nolan PM, Peters J, Strivens M, Rogers D, Hagan J, Spurr N, Gray IC, Vizor L, Brooker D, Whitehill E, Washbourne R, Hough T, Greenaway S, Hewitt M, Liu X, McCormack S, Pickford K, Selley R, Wells C, Tymowska-Lalanne Z, Roby P, Glenister P, Thornton C, Thaung C, Stevenson JA, Arkell R, Mburu P, Hardisty R, Kiernan A, Erven A, Steel KP, Voegeling S, Guenet JL, Nickols C, Sadri R, Nasse M, Isaacs A, Davies K, Browne M, Fisher EM, Martin J, Rastan S, Brown SD, Hunter J (2000) A systematic, genome-wide, phenotype-driven mutagenesis programme for gene function studies in the mouse. Nat Genet 25:440–443
Pettitt SJ, Liang Q, Rairdan XY, Moran JL, Prosser HM, Beier DR, Lloyd KC, Bradley A, Skarnes WC (2009) Agouti C57BL/6 N embryonic stem cells for mouse genetic resources. Nat Methods 6:493–495
Prosser HM, Koike-Yusa H, Cooper JD, Law FC, Bradley A (2011) A resource of vectors and ES cells for targeted deletion of microRNAs in mice. Nat Biotechnol 29:840–845
Ramírez-Solis R, Zheng H, Whiting J, Krumlauf R, Bradley A (1993) Hoxb-4 (Hox-2.6) mutant mice show homeotic transformation of a cervical vertebra and defects in the closure of the sternal rudiments. Cell 73:279–294
Ringwald M, Iyer V, Mason JC, Stone KR, Tadepally HD, Kadin JA, Bult CJ, Eppig JT, Oakley DJ, Briois S, Stupka E, Maselli V, Smedley D, Liu S, Hansen J, Baldock R, Hicks GG, Skarnes WC (2011) The IKMC web portal: a central point of entry to data and resources from the International Knockout Mouse Consortium. Nucleic Acids Res 39:D849–D855
Skarnes WC, Rosen B, West AP, Koutsourakis M, Bushell W, Iyer V, Mujica AO, Thomas M, Harrow J, Cox T, Jackson D, Severin J, Biggs P, Fu J, Nefedov M, de Jong PJ, Stewart AF, Bradley A (2011) A conditional knockout resource for the genome-wide study of mouse gene function. Nature 474:337–342
Stottmann RW, Beier DR (2010) Using ENU mutagenesis for phenotype-driven analysis of the mouse. Methods Enzymol 477:329–348
Stottmann RW, Moran JL, Turbe-Doan A, Driver E, Kelley M, Beier DR (2011) Focusing forward genetics: a tripartite ENU screen for neurodevelopmental mutations in the mouse. Genetics 188:615–624
Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC (1995) Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science 269:230–234
Wilkinson P, Sengerova J, Matteoni R, Chen CK, Soulat G, Ureta-Vidal A, Fessele S, Hagn M, Massimi M, Pickford K, Butler RH, Marschall S, Mallon AM, Pickard A, Raspa M, Scavizzi F, Fray M, Larrigaldie V, Leyritz J, Birney E, Tocchini-Valentini GP, Brown S, Herault Y, Montoliu L, de Angelis MH, Smedley D (2010) EMMA--mouse mutant resources for the international scientific community. Nucleic Acids Res 38:D570–D576
Acknowledgments
We thank members of the ICS unit and members of the EUCOMM (LSHG-CT-2005-018931), EUMODIC (LSHG-CT-2006-037188), Infrafrontier (FP7-INFRASTRUCTURES-2007-2.2.01/211404), INFRACOMP (FP7-INFRASTRUCTURES-2011-3.2/284501) EMMAService (FP7-INFRASTRUCTURES-2008-1/227490), and EUCOMMTools (LSHG-2011-261492) consortia for their helpful comments as participants of these programmes. We are grateful to the animal caretakers and technical staff who contributed to the two programmes as well as collaborators for useful discussion. This work was supported by the Wellcome Trust (grant number 098051), by the French state aid managed by the National Agency for Research under the program of future investments (ANR-10-INBS-07) and by several European funded projects listed above and especially EUMODIC (LSHG-CT-2006-560 037188).
Open Access
This article is distributed under the terms of the Creative Commons Attribution License which permits any use, distribution, and reproduction in any medium, provided the original author(s) and the source are credited.
Author information
Authors and Affiliations
Corresponding author
Additional information
A. Bradley, D. J. Adams, K. P. Steel, M. Hrabě de Angelis, S. D. Brown, and Y. Herault contributed equally to this study.
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 2.0 International License (https://creativecommons.org/licenses/by/2.0), which permits unrestricted use, distribution, and reproduction in any medium, provided the original work is properly cited.
About this article
Cite this article
Ayadi, A., Birling, MC., Bottomley, J. et al. Mouse large-scale phenotyping initiatives: overview of the European Mouse Disease Clinic (EUMODIC) and of the Wellcome Trust Sanger Institute Mouse Genetics Project. Mamm Genome 23, 600–610 (2012). https://doi.org/10.1007/s00335-012-9418-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-012-9418-y