Abstract
Recent linkage-based studies in humans suggest the presence of loci that affect either genome-wide recombination rates, utilization of recombination hotspots, or both. We have been interested in utilizing cytological methodology to directly assess recombination in mammalian meiocytes and to identify recombination-associated loci. In the present report we summarize studies in which we combined a cytological assay of recombination in mouse pachytene spermatocytes with QTL analyses to identify loci that contribute to genome-wide levels of recombination in male meiosis. Specifically, we analyzed MLH1 foci, a marker of crossovers, in 194 F2 male mice derived from a subspecific cross between CAST/EiJ and C57BL/6J parental strains. We then used these data to uncover loci associated with individual variation in mean MLH1 values. We identified seven recombination-associated loci across the genome (on chromosomes 2, 3, 4, 14, 15, 17, and X), indicating that there are multiple recombination “setting” loci in mammalian male meiosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Meiotic prophase is a complex process that requires coordinated activities of multiple protein families (e.g., cohesins, synaptonemal complex proteins, and recombination machinery proteins) to facilitate production of haploid gametes. The recombination events that occur between homologous chromosomes are especially important for normal chromosome disjunction. For example, in model organisms meiotic mutants that reduce recombination invariably increase meiotic nondisjunction (Marcon and Moens 2003; Ross-Macdonald and Roeder 1994; Yang et al. 2008). Similarly, analyses of humans indicate that abnormal numbers or placement of recombination events are important contributors to trisomies. Specifically, failure to recombine or crossovers located either too near to or too far from the centromere have been implicated in the origin of trisomies 13, 15, 16, 18, 21, and 22 and sex chromosome trisomies (Hassold et al. 2007). Nevertheless, despite the clinical importance of aberrant recombination, we remain remarkably ignorant of factors that influence the overall number of recombination events in oocytes or spermatocytes. Recombination rates are known to vary greatly across species and within species, among individuals and between sexes. For example, in humans genome-wide rates of recombination are approximately 1.6 times higher in females than in males (Matise et al. 2007) and can vary among individuals by as much as 20% (Cheng et al. 2009). However, relatively little is known about the factors that mediate this variation.
Similar to the situation in humans, we and others have noted strain-specific differences in recombination activity in mice. For example, recent high-resolution analyses of recombination on mouse chromosomes 1 and 17 have demonstrated differential usage of recombination hotspots among inbred strains (Baudat et al. 2010; Grey et al. 2009; Parvanov et al. 2009, 2010). Furthermore, in immunofluorescence-based studies that we conducted earlier this decade, we analyzed localization patterns of the DNA mismatch repair protein MLH1 in pachytene-stage spermatocytes of different inbred strains of mice. MLH1 is known to load onto pachytene-stage synaptonemal complexes at sites of crossovers (Baker et al. 1996; Froenicke et al. 2002; Koehler et al. 2002), and thus provides a useful marker for recombination events. In these studies we identified remarkable variation in mean MLH1 (crossover) counts per spermatocyte, with an approximate 15% difference between “low” CAST/EiJ (CAST) and “high” C57BL/6J (B6) recombination strains of mice. Accordingly, we recently exploited this variation to search for loci involved in setting recombination levels in mice. Specifically, as described in this report, we analyzed 194 F2 males derived from crosses between CAST and B6 animals, combining MLH1 assays of recombination with genome-wide QTL analyses to reveal loci associated with variation in recombination rates in male meiosis. We identified six autosomal loci and one X-linked locus, providing compelling evidence that multiple loci contribute to strain-specific variation in genome-wide recombination levels.
Materials and methods
Mouse information
Both mouse strains used in this study (CAST/EiJ and C57BL/6J) were obtained from The Jackson Laboratory (Bar Harbor, ME). Animals were housed in ventilated rack caging in a pathogen-free facility, with drinking water and feed provided ad libitum. C57BL/6J female mice were crossed with CAST/EiJ males to produce the F1 generation and F1 animals (brother × sister matings) were crossed at approximately 6 weeks of age. The F2 male offspring were used for cytological analysis at 6–8 weeks of age. All animal experiments were approved by the WSU Institutional Animal Care and Use Committee and conducted in accordance with the Guide for the Care and Use of Laboratory Animals. WSU is accredited by the American Association for Accreditation of Laboratory Animal Care.
Cytological analysis
Surface spread preparations from testicular samples were made using modifications of previously reported protocols (Peters et al. 1997) and the slides were immunostained with MLH1 and SYCP3 antibodies, respectively (Anderson et al. 1999; Koehler et al. 2002). The slides were examined on a Zeiss epifluorescence microscope, imaged with a CCD camera, and analyzed using Axiovision software. From each animal we identified pachytene-stage cells on the basis of the morphology of the synaptonemal complex (SC) (Moses 1980), and the total number of autosomal MLH1 foci per cell was determined for at least 25 cells. For each animal, cells were observed and counted by two independent observers.
Genotypic analysis
Tail snips were collected from each of the F2 males and stored at −20°C until all the sampling was finished. Genomic DNA was extracted from the tails using the Qiagen DNeasy kit (Qiagen, Valencia, CA) as per the manufacturer’s directions. The DNA was then quantified using a NanoDrop 2000 (NanoDrop Technologies, Wilmington, DE), and the concentrations of all the samples were standardized to 50 ng/μl. For the initial genome scan, genotyping was provided by the Center for Inherited Disease Research (CIDR) using their mouse medium-density linkage panel (Illumina, Inc., San Diego, CA), which contains 1449 single nucleotide polymorphisms (SNPs) across the genome at an approximate density of three SNPs per 5 Mb.
For analysis of allelic variation at Prdm9, genotypes were assessed based on the presence of sequence variation between B6 and CAST genomes. Standard PCR was performed using forward primer TGGTTTTATTGCTGTTGGCTTTC and reverse primer GTGCTCTTTGGCCTTCTCAAGT, designed to exon 12 of Prdm9 (XM_619431.4). The thermocycling conditions were as follows: 94°C for 2 min, followed by 34 cycles of 94°C for 30 s, 68°C for 30 s, 72°C for 1 min, with a final extension of 72°C for 5 min. Excess nucleotides and primers were cleaned from the PCR product using Exo SAP-IT® (USB Scientific, Affymetrix, Santa Clara, CA) as per the manufacturer’s recommended protocol. Sequencing reactions were performed using the reverse primer and BigDye® Terminator v3.1 with standard sequence conditions recommended by Applied Biosystems (Foster City, CA), with the exception of a higher (60°C) annealing temperature. Each sequence trace file was viewed with FinchTV (http://www.geospiza.com/Products/finchtv.shtml) software and genotyped based on sequence variation observed between the B6 and CAST genomes.
Data analysis
In initial analyses, we compared mean MLH1 values among B6, CAST, F1, and F2 animals (Fig. 1). However, for the subsequent QTL analyses, we used the square root of the average number of MLH1 foci per cell as the phenotypic variable. We first applied standard interval mapping (Lander and Botstein 1989). Genome-wide significance thresholds were calculated by a permutation test (Churchill and Doerge 1994), with a separate threshold for the X chromosome (Broman et al. 2006). We used 10,000 permutations for the autosomes and 136,400 permutations for the X chromosome. Subsequently, we performed multiple-QTL analyses using the stepwise selection algorithm and the penalized LOD score criterion of Manichaikul et al. (2009). All the QTL analyses were performed with R/qtl (Broman et al. 2003), and add-on package for the general statistical software R (R Development Core Team, 2010; http://www.r-project.org).
Results
Initially, we compared the genome-wide MLH1 values per cell among males of the two parental strains (CAST and B6; see Fig. 1). We analyzed 250 cells from 11 CAST males and 285 cells from 12 B6 males and confirmed our previously reported observations (Koehler et al. 2002), i.e., the mean number of MLH1 foci per cell for CAST males (21.4 ± 1.44) was significantly lower than the value for B6 males (24.0 ± 1.85); t = 5.78, p < 0.00001. Subsequently, we crossed the two strains, B6 × CAST, to produce F1 and F2 animals. The average MLH1 levels were determined for five F1 males and for the 194 F2 animals. These mean values were similar to one another and intermediate to the parental strains, i.e., for the F1s, 22.8 ± 1.83 (number of cells = 96) and for the F2s, 22.5 ± 1.82 (number of cells = 4845) (Fig. 1).
With the square root of the mean MLH1 values per cell as the phenotype, we then performed QTL analyses using all 194 F2 animals. Genotyping was provided by the Center for Inherited Disease Research (CIDR) using their mouse medium-density linkage panel (Illumina). This panel provided genotyping data for 732 informative single nucleotide polymorphism (SNP) markers, covering the genome at an approximate resolution of 2 cM. An initial genome scan revealed significant QTLs on chromosomes 2, 14, and X, with additional suggestive QTLs on chromosomes 3, 4, 15, and 17.
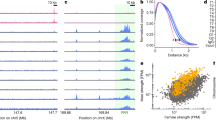
The fit and exploration of multiple-QTL models confirmed the associations to chromosomes 2, 3, 4, 14, 15, and X identified using the standard approach, and also indicated a significant association with a locus on chromosome 17. Profile LOD scores for all seven chromosomes are plotted in Fig. 2. The highest LOD score for each QTL, the chromosome location of the peak LOD, and the 1.5-LOD support interval of that region as well as the span that it encompasses are reported in Table 1. The X chromosome exhibited the highest LOD score (7.75), followed by chromosomes 2, 14, 3, 4, 17, and 15, respectively. The QTLs with the narrowest profiles, intervals of 13.0 cM, were on chromosomes 3 and 14. Conversely, the chromosome with the broadest profile, an interval of 30.3 cM, was on chromosome 4.
LOD profiles for a multiple-QTL model with loci on chromosomes 2, 3, 4, 14, 15, 17, and X. The LOD profile for a given chromosome is for a comparison between the 7-QTL model, with the position of the given QTLs allowed to vary but with the positions of the other 6 QTLs fixed at their estimated locations, and the 6-QTL model, with the given QTLs omitted. The dashed line denotes the position of the 5% estimated significance thresholds determined by permutation. The chromosomal locations of the peak LOD scores are shown in cM
Cumulatively, these loci explain an estimated 50% of the phenotypic variance, although this is likely an overestimate due to selection bias (Siegmund 2002). However, the direction of the effects of the loci varied. Specifically, the B6 allele was associated with higher recombination levels and the CAST allele with lower recombination levels on chromosomes 3, 4, 15, and 17, while the reverse situation applied to chromosomes 2, 14, and X. With the exception of chromosome 17, the effect of each allele was approximately additive (Fig. 3). We saw no evidence for epistatic interactions among the loci (data not shown).
Estimated phenotypic effects of individual QTLs. For chromosomes 3, 4, 15, and 17, the C57BL/6J allele (“B”) was associated with an increase in the number of MLH1 foci; for chromosomes 2, 14, and X, the CAST/Ei allele (“C”) was associated with an increase in MLH1 counts. For each chromosome, the relationship is shown for a specific SNP at the chromosomal location (in cM) yielding the highest LOD score. The vertical bars indicate ±1 SE
Subsequently, we examined the only gene (Prdm9) that has been linked to meiotic recombination activity in wild-type mice. Prdm9 is a histone methyltransferase-encoding gene that has been shown to affect utilization of recombination hotspots in different strains of mice. The Prdm9 locus is on chromosome 17, one of our candidate chromosomes, but is located just proximal to the 17-cM interval that we identified. Nevertheless, its strong effect on hotspot utilization prompted us to theorize whether it might be responsible for some of the variation between CAST and B6 males. Accordingly, we correlated allelic variation in Prdm9 with genome-wide MLH1 levels in the 194 F2 animals but we were unable to detect any obvious effect; indeed, the mean MLH1 values for animals with B6/B6, B6/CAST, or CAST/CAST genotypes were virtually identical to one another (data not shown).
Discussion
Until recently, most studies of loci involved in establishing meiotic recombination levels in mammals have relied on mutational analyses. For example, studies of mutant (typically knockout) mice have demonstrated the importance of SC formation, the establishment of sister chromatid cohesion, and DNA repair activities in promoting recombination, since null mutations in genes involved in each of these processes are associated with reduced or abolished crossing-over (Bolcun-Filas et al. 2007; Hunt and Hassold 2002; Kan et al. 2008). However, these advances notwithstanding, we still know little about loci controlling the number and placement of meiotic recombination sites in wild-type mammals or humans. Indeed, the first report of a gene important in determining recombination rates in humans came only 2 years ago when Kong et al. (2008) presented genetic linkage data from an Icelandic population correlating RNF212 with genome-wide recombination rates in humans. More recently, Chowdhury et al. (2009) confirmed the importance of RNF212 to male recombination levels and presented evidence of five other loci affecting recombination rates in either males or females.
In studies of recombination hotspots in mice, two groups recently reported a trans-regulator of recombination in the same chromosome region. Specifically, both Grey et al. (2009) and Parvanov et al. (2009) observed changes in hotspot usage in non-chromosome 17 regions in association with allelic variation in a region of chromosome 17. Subsequent analyses by these groups (Baudat et al. 2010; Parvanov et al. 2010) have implicated Prdm9, which is responsible for encoding a meiosis-specific protein responsible for trimethylation of histone H3K9. However, importantly, not all changes in recombination activity have been linked to chromosome 17 genotypes, indicating the existence of other, as yet unknown, recombination-associated loci.
In the present study we have taken a different approach to identification of recombination-setting loci, i.e., we combined a cytological assessment of recombination with standard QTL analysis to identify loci associated with genome-wide recombination levels. Our analyses indicate the presence of at least seven such QTLs. For four of these (on chromosomes 3, 4, 15, and 17), the associations are as one might expect, i.e., the allele originating from the parental strain with the highest levels of recombination (B6) was associated with an increase in average MLH1 localization in the F2 generation. In contrast, the results observed on chromosomes 2, 14, and X were somewhat surprising in that the allele originating from the parental strain with lower recombination rates (CAST) was associated with higher recombination levels in the F2 males. In fact, the average MLH1 localization was higher than that of the CAST parental strain, suggesting that these are transgressive QTLs.
Several intriguing candidate genes are housed in or near one of the seven intervals that we identified in the present study. Somewhat surprisingly, the mouse homolog of RNF212 is not one of these, as it is located on a chromosome (chromosome 5) not identified in our QTL analysis. Also unexpectedly, detailed genotypic analysis of one attractive candidate gene, Prdm9, located just proximal to our chromosome 17 QTL and the only gene known to affect recombination activity in mice, indicated no obvious effect on MLH1 values in the F2 animals. While the reason for this apparent discrepancy is not immediately obvious, we can think of at least two possible explanations. First, previous studies of Prdm9 in mice have focused on hotspot utilization, and it seems unlikely that there is a simple, direct relationship between this aspect of recombination activity and genome-wide MLH1 rates (e.g., references in Parvanov et al. 2009). Alternatively, it may well be that there is a relationship between Prdm9 genotype and MLH1 values, but it is below the level of detection afforded by our present study population. Studies to distinguish between these or other possible explanations are in progress.
Of the other QTLs, the most significant LOD score was associated with an interval on the X chromosome, an observation consistent with other reports of X-linked recombination-associated loci (de La Casa-Esperon et al. 2002; Ng et al. 2009). A gene of interest in close proximity to our most significant associated X-linked SNP is testis-expressed gene 11 (Tex11), first identified as an X-linked germ cell-specific gene by Wang et al. (2001) and later reported to be essential for male meiosis (Yang et al. 2008; Zheng et al. 2010). TEX11 is thought to interact with SYCP2 to promote synapsis and crossover formation (ref) and thus is a strong candidate for a recombination-setting locus. Studies designed to test the importance of TEX11 to male recombination are presently underway.
References
Anderson LK, Reeves A, Webb LM, Ashley T (1999) Distribution of crossing over on mouse synaptonemal complexes using immunofluorescent localization of MLH1 protein. Genetics 151:1569–1579
Baker SM, Plug AW, Prolla TA, Bronner CE, Harris AC et al (1996) Involvement of mouse Mlh1 in DNA mismatch repair and meiotic crossing over. Nat Genet 13:336–342
Baudat F, Buard J, Grey C, Fledel-Alon A, Ober C et al (2010) PRDM9 is a major determinant of meiotic recombination hotspots in humans and mice. Science 327:836–840
Bolcun-Filas E, Costa Y, Speed R, Taggart M, Benavente R et al (2007) SYCE2 is required for synaptonemal complex assembly, double strand break repair, and homologous recombination. J Cell Biol 176:741–747
Broman KW, Wu H, Sen S, Churchill GA (2003) R/qtl: QTL mapping in experimental crosses. Bioinformatics 19:889–890
Broman KW, Sen S, Owens SE, Manichaikul A, Southard-Smith EM et al (2006) The X chromosome in quantitative trait locus mapping. Genetics 174:2151–2158
Cheng EY, Hunt PA, Naluai-Cecchini TA, Fligner CL, Fujimoto VY et al (2009) Meiotic recombination in human oocytes. PLoS Genet 5:e1000661
Chowdhury R, Bois PR, Feingold E, Sherman SL, Cheung VG (2009) Genetic analysis of variation in human meiotic recombination. PLoS Genet 5:e1000648
Churchill GA, Doerge RW (1994) Empirical threshold values for quantitative trait mapping. Genetics 138:963–971
de La Casa-Esperon E, Loredo-Osti JC, Pardo-Manuel de Villena F, Briscoe TL, Malette JM et al (2002) X chromosome effect on maternal recombination and meiotic drive in the mouse. Genetics 161:1651–1659
Froenicke L, Anderson LK, Wienberg J, Ashley T (2002) Male mouse recombination maps for each autosome identified by chromosome painting. Am J Hum Genet 71:1353–1368
Grey C, Baudat F, de Massy B (2009) Genome-wide control of the distribution of meiotic recombination. PLoS Biol 7:e35
Hassold T, Hall H, Hunt P (2007) The origin of human aneuploidy: where we have been, where we are going. Hum Mol Genet 16(Spec No. 2):R203–R208
Hunt PA, Hassold TJ (2002) Sex matters in meiosis. Science 296:2181–2183
Kan R, Sun X, Kolas NK, Avdievich E, Kneitz B et al (2008) Comparative analysis of meiotic progression in female mice bearing mutations in genes of the DNA mismatch repair pathway. Biol Reprod 78:462–471
Koehler KE, Cherry JP, Lynn A, Hunt PA, Hassold TJ (2002) Genetic control of mammalian meiotic recombination. I. Variation in exchange frequencies among males from inbred mouse strains. Genetics 162:297–306
Kong A, Thorleifsson G, Stefansson H, Masson G, Helgason A et al (2008) Sequence variants in the RNF212 gene associate with genome-wide recombination rate. Science 319:1398–1401
Lander ES, Botstein D (1989) Mapping mendelian factors underlying quantitative traits using RFLP linkage maps. Genetics 121:185–199
Manichaikul A, Moon JY, Sen S, Yandell BS, Broman KW (2009) A model selection approach for the identification of quantitative trait loci in experimental crosses, allowing epistasis. Genetics 181:1077–1086
Marcon E, Moens P (2003) MLH1p and MLH3p localize to precociously induced chiasmata of okadaic-acid-treated mouse spermatocytes. Genetics 165:2283–2287
Matise TC, Chen F, Chen W, De La Vega FM, Hansen M et al (2007) A second-generation combined linkage physical map of the human genome. Genome Res 17:1783–1786
Moses MJ (ed) (1980) New cytogenetic studies on mammalian meiosis. Raven Press, New York
Ng SH, Madeira R, Parvanov ED, Petros LM, Petkov PM et al (2009) Parental origin of chromosomes influences crossover activity within the Kcnq1 transcriptionally imprinted domain of Mus musculus. BMC Mol Biol 10:43
Parvanov ED, Ng SH, Petkov PM, Paigen K (2009) Trans-regulation of mouse meiotic recombination hotspots by Rcr1. PLoS Biol 7:e36
Parvanov ED, Petkov PM, Paigen K (2010) Prdm9 controls activation of mammalian recombination hotspots. Science 327:835
Peters AH, Plug AW, van Vugt MJ, de Boer P (1997) A drying-down technique for the spreading of mammalian meiocytes from the male and female germline. Chromosome Res 5:66–68
Ross-Macdonald P, Roeder GS (1994) Mutation of a meiosis-specific MutS homolog decreases crossing over but not mismatch correction. Cell 79:1069–1080
Siegmund D (2002) Upward bias in estimation of genetic effects. Am J Hum Genet 71:1183–1188
Wang PJ, McCarrey JR, Yang F, Page DC (2001) An abundance of X-linked genes expressed in spermatogonia. Nat Genet 27:422–426
Yang F, Gell K, van der Heijden GW, Eckardt S, Leu NA et al (2008) Meiotic failure in male mice lacking an X-linked factor. Genes Dev 22:682–691
Zheng K, Yang F, Wang PJ (2010) Regulation of male fertility by X-linked genes. J Androl 31:79–85
Acknowledgment
This research was supported by NIH grants HD21341 and HD42720 to T. Hassold and GM074244 to K. Broman.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Murdoch, B., Owen, N., Shirley, S. et al. Multiple loci contribute to genome-wide recombination levels in male mice. Mamm Genome 21, 550–555 (2010). https://doi.org/10.1007/s00335-010-9303-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-010-9303-5