Abstract
The adrenomedullary hormone epinephrine transduces environmental stressors into cardiovascular events (tachycardia and hypertension). Although the epinephrine biosynthetic enzyme PNMT genetic locus displays both linkage and association to such traits, genetic variation underlying these quantitative phenotypes is not established. Using an integrated suite of computational and experimental approaches, we elucidate a functional mechanism for common (minor allele frequencies > 30%) genetic variants at PNMT. Transcription factor binding motif prediction on mammalian PNMT promoter alignments identified two variant regulatory motifs, SP1 and EGR1, disrupted by G-367A (rs3764351), and SOX17 motif created by G-161A (rs876493). Electrophoretic mobility shifts of approximately 30-bp oligonucleotides containing ancestral versus variant alleles validated the computational hypothesis. Queried against chromaffin cell nuclear protein extracts, only the G-367 and -161A alleles shifted. Specific antibodies applied in electrophoretic gel shift experiments confirmed binding of SP1 and EGR1 to G-367 and SOX17 to -161A. The in vitro allele-specific binding was verified in cella through promoter reporter assays: lower activity for -367A haplotypes cotransfected by SP1 (p = 0.002) and EGR1 (p = 0.034); and enhanced inhibition of -161A haplotypes (p = 0.0003) cotransfected with SP1 + SOX17. Finally, we probed cis/trans regulation with endogenous factors by chromatin immunoprecipitation using SP1/EGR1/SOX17 antibodies. We describe the systematic application of complementary computational and experimental techniques to detect and document functional genetic variation in a trait-associated regulatory region. The results provide insight into cis and trans transcriptional mechanisms whereby common variation at PNMT can give rise to quantitative changes in human physiological and disease traits. Thus, PNMT variants in cis may interact with nuclear factors in trans to govern adrenergic activity.
Similar content being viewed by others
Introduction
The enzyme PNMT (phenylethanolamine N-methyltransferase) catalyzes the synthesis of epinephrine (adrenaline), providing the only route to formation of this hormone and neurotransmitter. Variation in the human PNMT promoter was first noted by Wu and Comings (1999) and later systematically documented in European-American, African-American, and Japanese populations (Ji et al. 2005; Saito et al. 2001). Two common PNMT proximal promoter SNPs are located -367 (rs876493) and -161 (rs3764351) bp upstream of the 5′ UTR (untranslated region) (Kaneda et al. 1988), corresponding to -390 and -184 bp upstream of the translational start (ATG) codon (Sasaoka et al. 1989). Reported linkage disequilibrium (LD) is high (D′ > 0.9) across the promoter in the HapMap CEU population (Utah residents with Northern and Western European ancestry from the Centre d'Etude du Polymorphisme Humain collection) (Frazer et al. 2007; Thorisson et al. 2005).
The Pnmt locus is within the genome-wide confidence interval for linkage (meiotic cosegregation) with several stress traits in rodents (http://rgd.mcw.edu/), including salt-loaded SBP QTL-9, urine volume QTL-18, and corticosterone (glucocorticoid) level QTL-5. Several human disease associations to PNMT promoter genetic variants have been documented, including hypertension in African-Americans (Cui et al. 2003), early-onset Alzheimer’s disease (Mann et al. 2001), multiple sclerosis (Mann et al. 2002), drug-assisted weight loss (Peters et al. 2003), and reward dependence temperament (Yamano et al. 2008). Although two studies failed to find association with hypertension in European (Kepp et al. 2007) and European-American (Cui et al. 2003) subjects, we observed sex-dependent association to diastolic blood pressure (Rana et al. 2007).
Prior PNMT genetic studies have not definitively identified molecular mechanisms whereby genetic variation may interact with stressors to change PNMT expression. Ji et al. (2005) compared PNMT promoter haplotype activity, observing a significant decrease in activity for the A.A (-367.-161) haplotype versus G.G. Extensive work in the PC12 rat pheochromocytoma cell line suggests orthologous regulatory motifs for SP1, EGR1, AP2, MAZ, and GRE in the first 1000 base pairs of proximal promoter (Wong and Tank 2007).
Here we explore regulatory effects of common PNMT promoter genetic variants (G-367A, G-161A) in cis, as well as the effects of factors in trans, including SP1 (at G-367A) and SOX17 (at G-161A), via orthologous motifs in the human promoter, demonstrating an allele-specific effect for SP1 and EGR1 on the G-allele at position-367. Furthermore, we identify a novel allele-specific inhibitory effect of SOX17 interacting with SP1 to preferentially inhibit the A allele at position -161. Sox17 is a transcription factor involved in development of heart and vascular and lung tissue (Liao et al. 2009; Liu et al. 2007; Matsui et al. 2006; Park et al. 2006); all are important targets of the epinephrine-mediated stress response.
Materials and methods
Genomics and luciferase promoter/reporter plasmids
Human PNMT sequence information was obtained at NCBI (http://www.ncbi.nlm.nih.gov) or UCSC ( http://genome.ucsc.edu) using source clone X52730 (JO3727) (Sasaoka et al. 1989). Mammalian promoter sequencing and polymorphism discovery at the PNMT locus in genomic DNA were conducted using dideoxy sequencing on an ABI 3100 capillary sequencer (Applied Biosystems, Foster City, CA). The PNMT promoter regions in chimpanzee (NG06939), bonobo (NG05253), and rhesus (NG07109) DNA from Coriell repository samples, as well as rat (strains SHR, WKY, and BN) samples, were resequenced.
At PNMT, two promoter variants (G-367A and G-161A) were especially common minor allele frequency >30% (Ji et al. 2005). The very proximal core promoter (Fig. 1) just upstream of exon 1, including the TATA box, was invariant; there were also six G/C-rich stretches, one of which was disrupted by G-367A. The two common promoter variants (-367.-161) displayed consistent LD, with D′ approximately 0.90 in Caucasians. Of the four possible promoter haplotypes (Ji et al. 2005) at -367.-161, the A.A (at 50.6%) and G.G (35.1%) were common, while A.G (12.0%) and G.A (1.9%) were unusual. Based on promoter location, high frequencies, and LD of promoter variants G-367A and G-161A, we then focused functional studies on this region.
Human PNMT promoter/luciferase reporter plasmids were constructed as previously described (Chen et al. 2008). G.G and A.G haplotype fragments corresponding to PNMT −428 to +10 bp [with respect to cap site of 23 bp 5′ UTR in NCBI source clone X52730 (J03727): 1327-1793, Chr 17: 35077803-35078242] were amplified from genomic DNA of known (previously resequenced) promoter genotype (G.G, 68-year-old black female [#2154]; A.G, 45-year-old Hispanic male [#4261]); these two individuals did not exhibit other PNMT promoter variants. The haplotype fragments were subcloned by sticky-end ligation into the upstream promoter site of the pGL3-Basic vector (Promega Inc., Madison, WI, USA) between XhoI and KpnI restriction digest sites (New England Biolabs, Ipswich, MA, USA). Successful ligations were verified by electrophoresis on 1% agarose of 1 μg plasmid digested overnight with KpnI and XhoI; correct sequence and orientation were verified by sequencing with RV3 reverse primer (Promega). G.A and A.A haplotypes (-367.-161) were generated by site-directed mutagenesis per the manufacturer’s specifications (QuickChange II; Stratagene, La Jolla, CA, USA) using the aforementioned G.G and A.G haplotype plasmids as template and the same oligonucleotide primers as in the electrophoretic mobility shift assay (EMSA) experiments.
Plasmids were amplified in DH5-alpha cells (Invitrogen, Carlsbad, CA, USA), and supercoiled plasmids were purified by column adsorption/elution (PureLink HiPure Plasmid Maxiprep Kit, Invitrogen). Plasmid concentrations were quantified by SmartSpec (Bio-Rad, Hercules, CA, USA) and adjusted to 1.0 μg/μl before transfection.
Cell biology: transient promoter haplotype transfection and reporter assay
PC12 rat adrenal pheochromocytoma cells were grown in high-glucose Dulbecco’s modified Eagle medium supplemented with penicillin G (100 U/ml), streptomycin sulfate (100 mg/ml), 5% fetal bovine serum, and 10% horse serum. PC12 cells were distributed among 12-well plates and allowed to reach 60% confluence before transfection. Plasmid DNA was incubated in 100 μl per well of serum-free media with TransfectIN reagent (Bio-Rad) for 15 min and then added to 400 μl fresh PC12 media. Cells were transiently transfected for 4 h with 1 μg per well haplotype plasmid or pGL3-Basic vector and then washed with serum-free media followed by 1 ml PC12 media. Firefly luciferase activity was measured after 48 h of incubation at 37°C, 6% CO2. Luciferase activity was normalized by total protein per well measured by Bradford assay (Protein Assay, Bio-Rad) in a Wallac spectrophotometer. Linear range of protein concentration was verified using serial dilutions of bovine serum albumin (BSA) as a standard. pCMV-driven transcription factor expressing plasmids, including EGR1, AP2, SOX17, and SP1 plasmids (from Open Biosystems, Huntsville, AL, USA), were cotransfected at 0.1 μg per well. Dual cotransfection of SP1 with SOX17 or SP1 with pcDNA (pCMV) was also tested at 0.1 μg per well.
PNMT promoter haplotype/luciferase reporter activity
Haplotype luciferase activity (RLU) was normalized to total protein (Bio-Rad/Bradford Coomassie blue assay at A595; also background subtracted and divided by volume). The activity shown in Fig. 3 is the mean activity for four wells ±1 standard error. In order to compare experiments, we normalize all values to the empty vector pGL3-Basic activity. Finally, in order to control for effects of plasmid cotransfection on promoter activity, we subtract the activity of the mock cotransfection of the pcDNA3.1 plasmid, which contains the promoter used to drive transcription factor expression.
Bioinformatics
Mammalian promoter alignment
Mouse, dog, cat, horse, and cow PNMT sequences were obtained from http://genome.ucsc.edu (Blanchette et al. 2004; Kent et al. 2002; Kuhn et al. 2009). Due to a gap in the R. norvegicus reference genome assembly, we also sequenced the promoter of the WKY rat. Promoter orientation for each sequence was determined by ClustalW alignment of the exon 1 coding sequence. Promoter sequences were aligned using ClustalW with a gap penalty of 100. Phylogenetic trees of the mammals were constructed manually using taxonomy information from NCBI.
Prediction of allele-specific conserved regulatory motifs
We predicted transcription factors with allele-specific binding to motifs containing G-367 or G-161A using phylogenetic footprinting (Wasserman and Sandelin 2004). Human and rat PNMT promoter sequences were pairwise aligned using the LAGAN algorithm (Brudno et al. 2003), as implemented by the CONREAL web server (Berezikov et al. 2005). Motifs with more than 50% human-rat homology (length > 10 bp) were scored on publicly available position-weight matrices (PWMs) from JASPAR (Vlieghe et al. 2006) and TRANSFAC v7.0 (Wingender et al. 1996) public databases. Motifs scoring greater than 80% of maximum in one allele with a difference in score between alleles were considered positive hits (summarized in Fig. 2 using SOX17 and SP1 as examples). Results were cross-verified using the RAVEN web server (Andersen et al. 2008), which uses seven-vertebrate PhastCons (Felsenstein and Churchill 1996) alignments for identification of conserved motifs. Candidate hits were prioritized for experimental validation if known to regulate the well-characterized rat PNMT promoter (Wong and Tank 2007), or if adrenal, chromaffin cell, or PC12 expression activity in GEO (Edgar et al. 2002) expression array repository was above 25% of minimum.
Allele- and factor-specific transcription enhancement and inhibition in the human PNMT promoter. ClustalW alignment of orthologous mammalian PNMT promoter motifs, including two human alleles, three primates, and six other mammals, with (a) -367 and (b) -161 positions in lowercase. Rat-human alignments shown with greater than 50% conserved motif in uppercase for (c) -367 and (d) -161. JASPAR position frequency matrix scoring of human sequence (boxes) using (e) SP1 on -367 and (f) SOX17 on -161. Polymorphic position in boldface type. Using the SOX17 JASPAR motif (YWYTGKB), there are an additional 22 nonpolymorphic SOX17 sequence matches at 7 of 8 or better stringency in the first 500 bp of proximal promoter. Using a conventional G/C-rich motif (Sn) consensus, there are an additional five nonpolymorphic matches spanning 11–18 bp each (see Fig. 1)
Electrophoretic mobility shift assay (EMSA)
Biotinylated double-stranded 29-mer oligonucleotides (ValueGene, Biotin 3′ End DNA Labeling Kit, Pierce, Rockford, IL, USA) with SNP at position 15 were incubated 45 min and with nuclear extracts of PC12 rat pheochromocytoma cells for 20 min in the presence of 0.1 μg/μl salmon sperm DNA to minimize nonspecific binding. Nuclear extracts were prepared from PC12 cells grown in 10-cm plates by Qproteome Nuclear Protein Kit following the manufacturer’s specifications (Qiagen) (Avery-Kiejda et al. 2008). In separate reactions, either specific (i.e., unlabeled same-allele oligonucleotides) or nonspecific competitor (i.e., unlabeled opposite-allele oligonucleotides) was added at 1000× concentration to assay allele-specific binding. Antibody assays included 2 μl of antibody (Santa Cruz Biotechnology, Santa Cruz, CA, USA) in lieu of competitor oligonucleotide, with nonspecific IgG as a negative control). DNA-protein complexes were electrophoresed in 5% acrylamide gels for 1 h at 100 V in 0.5× tris-borate ethylene diamine tetra acetic acid (TBE) and then transferred to nylon membranes at 380 mA in 0.5× TBE and cross-linked with 1200 nm UV light for 1 min. Complexes were then visualized on X-ray film using horseradish-peroxidase-conjugated streptavidin per the manufacturer’s protocol (Chemiluminescent Nucleic Acid Detection Module, Pierce, Rockford, IL, USA) (Matata and Galinanes 2002; Oltra et al. 2003). Forward/reverse allele-specific oligonucleotide probes were as follows (polymorphism in lowercase):
-
-367 A fw: 5′-TGAATGGCTGCGGGaGGCTGGAGAAGAGA-3′
-
-367 A rv: 5′-TCTCTTCTCCAGCCtCCCGCAGCCATTCA-3′
-
-367 G fw: 5′-TGAATGGCTGCGGGgGGCTGGAGAAGAGA-3′
-
-367 G rv: 5′-TCTCTTCTCCAGCCcCCCGCAGCCATTCA-3′
-
-161 G fw: 5′-CCCCACTCACCTCCgGTGTGTCTGCAGCC-3′
-
-161 G rv: 5′-GGCTGCAGACACACcGGAGGTGAGTGGGG-3′
-
-161 A fw: 5′-CCCCACTCACCTCCaGTGTGTCTGCAGCC-3′
-
-161 A rv: 5′-GGCTGCAGACACACtGGAGGTGAGTGGGG-3′
Chromatin ImmunoPrecipitation (ChIP)
PC12 cells at 70% confluence grown on 10-cm poly-l-lysine-coated plates were transiently transfected with 5 μg promoter haplotype/reporter plasmid DNA 4 h (Bio-Rad transfectin). Cells were washed with serum-free medium (DMEM high-glucose), grown 24 h (5% FBS, 10% HS, 1% PSG), washed with phosphate-buffered saline (PBS), fixed with 37% formaldehyde, pelleted, and transferred to lysis buffer for fragmentation of chromatin to nucleosomes by controlled sonication (11 cycles in a Branson Sonifier; 20-s pulse at 50% amplitude with 1 min cooling). Nucleosomes were immunoprecipitated with antibodies for EGR1 (Santa Cruz Biotechnologies; sc-110x), SP1 (sc-420x), SOX17 (sc-20099x), or IgG/pre-immune serum (“Mock”; sc-33608) conjugated to Protein A agarose beads (ChIP Assay Kit, USB Corporation, Cleveland, OH, USA). After precipitation, complexes were washed and crosslinking was reversed at 65°C overnight. After column purification (QiaQuick PCR Purification, Qiagen), DNA was amplified for 26–30 cycles (to preclude signal from the negative control) with SNP-flanking primers designed using PRIMER3, and products were electrophoresed on 1.5% agarose gels and photographed with a digital camera. Primers were as follows:
-
G-367A fw: 5′-CTGGGCAAAAGAGGTGAATG-3′
-
G-367A rv: 5′-CCTCACCCCCACAATCTTTA-3′
-
G-161A fw: 5′-CTGCGGGAGCAAGTACGG-3′
-
G-161A rv: 5′-CCCACCCATTCATCCATCT-3′
Statistics
Changes in promoter haplotype/reporter activity were evaluated by one-way or two-way analysis of variance (ANOVA), as appropriate, using SPSS software (SPSS, Inc., Chicago, IL, USA), using (as the independent variable) either the haplotype or the SNP. Post-hoc corrections to significance were done by the Fisher LSD (least significance difference) method.
Results
SNP-interrupted conserved regulatory motifs in the PNMT promoter
Alignment of orthologous 1-kb promoters (upstream of the ATG initiation codon), including three primates, six mammals, and human reference/variant sequences, showed a moderately conserved G/C-rich (GGGGGG) motif in primates, interrupted by the A allele at position -367 in the human promoter (GCGGG[G/A]GGC; Fig. 2a). In chimpanzee, bonobo, and rhesus, G occurs at the polymorphic site; however, in the less conserved region in mouse, rat, dog, cat, horse, and cow, the allele is A.
At human promoter position -161, G is the major primate allele (Fig. 2b); however, the surrounding bases are not well conserved with rodents and nonprimate mammals. This region of the promoter is divergent across mammals but conserved within subgroups as clustered by the phylogenetic tree in Fig. 2 (mouse and rat, dog and cat, horse and cow all have pairwise-conserved motifs).
Phylogenetic footprinting was used to predict which transcription factors are responsible for the allele-specific EMSA shifts observed in Fig. 4. Alignments with rat promoter for human sequences flanking promoter positions -367 (Fig. 2c, human G allele shown) and -161A (Fig. 2d, human A allele shown) identified regions with greater than 50% sequence identity (in uppercase). Position frequency matrix scoring using JASPAR matrices (SP1 on -367 G vs. A, Fig. 2e; SOX17 on -161 G vs. A, Fig. 2f) highlights allelic differences: at G-367A, a two-point difference between alleles, with the -367 G allele scoring higher for SP1 (88% of maximum score), and at G-161A there was a 30-point difference between alleles, with -161 A scoring higher on SOX17 (TCC[G/A]GTGTG, 80% of maximum score). Scoring matrices were selected from the top hits identified using CONREAL, with both SP1 and SOX17 mRNA expression in PC12 cells more than 50% of maximum in GEO profiles of PC12 cells.
Allele-specific transcription: functional enhancement or inhibition of expression
The transcription factors tested in cotransfection assays (Fig. 3) were selected by a combination of in silico and in vitro predictions of regulatory function; at G-367A, G/C-rich-region candidates SP1 and EGR1 were tested, while at G-161A, factor SOX17 was evaluated. We compared the transcriptional response of four PNMT promoter haplotypes (G-367A.G-161A, illustrated in Fig. 3a) to transcription factors EGR1, SP1, and SOX17. Even though haplotype G.A did not occur in human samples, creation of this fourth haplotype (by site-directed mutagenesis) allowed statistical inferences on each of the two contributory SNPs, by two-way ANOVA. Graphical and statistical results are shown in Fig. 3.
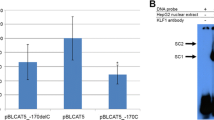
SNP-interrupted conserved regulatory motifs in the human PNMT promoter: functional consequences. a Cartoon of PNMT promoter/reporter construct, one of four possible promoters based on variation at positions G-367A/G-161A (G.A, A.G, A.A, G.G), which were ligated 5′ of firefly luciferase in pGL3-Basic plasmid and transfected into PC12 cells. b Basal (unstimulated) promoter activity for transfected PNMT promoter haplotypes (G-367A/G-161A; G.A, A.G, A.A, G.G), measured in cell lysates after 48 h of transient transfection, mean of four wells ± 1 standard error. Statistics: Haplotype effect (1-way ANOVA, overall p = 0.009) is shown first. Two-way ANOVA is then shown, factoring by each SNP. Horizontal bars indicate post-hoc t tests between haplotypes. X axis: Promoter haplotype; Y axis: Promoter activity in RLU/protein. c Change in promoter activity for PNMT promoter haplotypes, measured in cell lysates after 48 h of transient transfection; mean of four wells ± 1 standard error. Y axis shows fold change of activity versus pGL3-Basic promotorless empty vector, with mock pcDNA3.1 (pCMV) cotransfection activity subtracted, and normalization by total protein (quantified by the Bradford assay). Along the X axis are grouped the transfected promoter haplotypes (G-367A.G-161A) and the pCMV-driven transcription factors cotransfected (SP1, EGR1, SOX17, and SP1 + SOX17). Horizontal bars indicate post-hoc t tests for haplotype comparisons within a cotransfection state. d ANOVA p values for overall effect, individual SNP effects, and SNP × SNP (G-367A × G-161A) interactions, during transcriptional activation by cotransfection
Basal haplotype expression
Under basal circumstances, the four haplotypes differed in transcriptional strength (Fig. 3b) overall (ANOVA p = 0.009), with a significant effect of G-367A (p = 0.002), but without SNP × SNP interaction (two-way ANOVA p = 0.163). As previously reported (Ji et al. 2005), haplotype G.G was more active than haplotype A.A (p = 0.009).
G-367A and SP1 or EGR1
We observed a significant difference in promoter stimulation (Fig. 3d) between G-367A alleles upon SP1 cotransfection (p = 0.002), with higher activity for the G allele that displayed a superior SP1 match (Fig. 3c). The response of the G.A haplotype to SP1 was greater (p = 0.005) than either of the A.X haplotypes (A.G or A.A). EGR1 trans activation also differed across PNMT promoter haplotypes (p = 0.035).
G-161A and SOX17
SOX17 cotransfection produced a modest decline (p = 0.014) in the activity of each variant haplotype except G.A (Fig. 3c); there were significant differences between G-161A alleles (p = 0.029) and G-367A alleles (p = 0.008) (Fig. 3d).
Factor cotransfection
In order to further characterize the transcription inhibition effect of SOX17 on the PNMT promoter, we attempted dual/cotransfection of SP1 and SOX17. Overall, we observed an enhanced, supra-additive inhibitory effect upon transcription (p = 0.003) (Fig. 3c, far right), especially on G.A and A.A haplotypic backgrounds. We also observed an allelic difference at G-161A in response to the factors in combination (SOX17 + SP1, p = 0.0003), with lower activity for the A allele.
Allele-specific factor binding: EMSAs and specific antibody disruption
The work described in Figs. 2, 3, 4 reflects a cyclical in silico → in vitro → in cella approach to dissecting allele-specific transcription regulation by specific transcription factors. Allele-specific shifts of human oligonucleotides bound to rat chromaffin cell nuclear proteins observed in Fig. 4 prompted the screening for position-weight matrices scored in Fig. 2. The regulatory effect of these factors is confirmed in vivo in Fig. 3. However, regulatory stimulation does not guarantee binding of the putative transcription factor to a specific motif. Hence, we select antibodies based on the combined results to confirm allele-specific binding in vitro in an EMSA antibody disruption assay (Fig. 5).
Allele-specific electrophoretic mobility shifts. Electrophoresis at 90 V of double-stranded biotinylated oligonucleotides incubating in the presence/absence of PC12 nuclear DNA-binding protein extracts (water-soluble) and 1000 × molar excess unlabeled oligonucleotide. Visualization of bands by trans-blot to nylon membrane, streptavidin-horseradish-peroxidase conjugation, and incubation for 5 min with luminol solution, followed by exposure on X-ray film. Horizontal black bar separates G- (left) from A- (right) allele oligonucleotide experiments (label on top of image). Oligonucleotide sequences designed to match 29-bp motifs at a -367 and b -161, with polymorphic position in brackets (bottom). Center bar illustrates migration from top to bottom and highlights G-allele-specific shift for -367 oligonucleotide and A-allele-specific shift for -161 oligonucletotide
Electrophoretic mobility shifts: Perturbation by transcription factor-specific antibodies. Electrophoresis at 90 V of double-stranded biotinylated oligonucleotides incubate in the presence/absence of PC12 nuclear DNA-binding protein extracts (water-soluble) and antibodies to a SP1 and EGR1 versus -367 oligonucleotides or b anti-SOX17 and anti-SP1 versus -161 oligonucleotides. No-antibody shift same as Fig. 4 used as a positive control. Nonspecific (preimmune) IgG (“Mock”) was a negative control. Same protocol as Fig. 4
During EMSA of G-367A (Fig. 4a), the labeled G allele displayed multiple (2–3) shifts by PC12 nuclear proteins, which could be partially displaced by the unlabeled G probe, although not the unlabeled A probe. The labeled A allele was not shifted by nuclear proteins, indicating specificity. Based on the phylogenetic footprinting results shown in Fig. 2, we tested the effect of antibodies to SP1 (Fig. 5a, lane 3) or EGR1 (lane 4); in each case, the shifted G-allele bands (lane 2) were disrupted or abolished by coincubation with the antisera, suggesting involvement of these two factors. Involvement of EGR1 at G-367A is further illustrated by Supplementary Fig. 1, pointing out the overlapping EGR1 motif match, which is superior for the G allele.
During EMSA of G-161A, we observed multiple electrophoretic mobility shift bands for both labeled alleles (Fig. 4b, lanes 2 and 6). In each case, the labeled allele shift is best competed for by the unlabeled A allele (Fig. 4b, lanes 4 and 7), suggesting higher affinity of the A allele for the nuclear proteins bound. Based on phylogenetic footprinting results shown in Fig. 2, we tested the effects of an anti-SOX17 antibody on the complex (Fig. 5b, lane 9); this addition also manifested as a disappearance of the previously shifted A-allele band (lane 9), once again suggesting antibody sequestration of nuclear SOX17, freeing the SOX17 binding site on the -367A allele oligonucleotide. We did not observe such effects at G-161A using SP1 or mock/IgG antibodies, confirming specificity (Fig. 5b, lanes 8 and 10). Furthermore, what appears to be a G-allele shift (Fig. 5b, lane 2; Fig. 5b, lane 2) was not displaced by any of the tested antibodies, and hence is of uncertain significance.
Haplotype-specific transcription factor binding to the motifs in chromatin
Formation of oligonucleotide-factor-antibody complexes in vitro is a model but not a substitute for in vivo complex formation in the cell nucleus. Although the results of the cotransfection promoter/reporter assays, phylogenetic footprinting, and electrophoretic mobility shift assays produce a consistent story of SP1 and EGR1 upregulation on the G allele at human promoter position -367 and of SOX17 downregulation on the A allele at position -161, we attempted to extend this result by chromatin immunoprecipitation (ChIP), which may yield insight into endogenous transcription factor binding by a motif in nuclei prepared from living cells.
Antibodies to both SP1 and EGR1 immunoprecipitated nucleosomes harboring the proximal PNMT promoter (Fig. 6b), as evidenced by the PCR amplicon spanning G-367A; specificity was indicated by lack of immunoprecipitation by preimmune serum (“Mock”). Thus, endogenous SP1 and EGR1 seem to interact with the PNMT proximal promoter within chromatin in vivo, although the results of this semiquantitative experiment did not distinguish differential roles for the G and A alleles.
Haplotype-specific endogenous transcription factor binding in the human PNMT promoter. Chromatin ImmunoPrecipitation (ChIP) assay to determine binding of particular transcription factors to the motifs in vivo. a Cartoon of primer design and expected PCR amplification fragment size. Primers specific for amplification of b -367 were used to identify DNA-protein complexes precipitated with anti-EGR1, anti-SOX17, or nonspecific IgG (“Mock,” preimmune serum) antibodies, or raw sonicated DNA (“Input”) as a positive control. c Identical experiment testing anti-SOX17 with -161 primers
An antibody to SOX17 also immunoprecipitated nucleosomes spanning the PNMT promoter (Fig. 6c), as evidenced by the PCR amplicon spanning G-161A, although the G and A alleles did not differ in band intensity; specificity was indicated by lack of immunoprecipitation by preimmune serum (“Mock”). Once again, lack of difference for the G and A alleles may simply reflect the semiquantitative nature of the experiment.
Discussion
Through an integrated in silico → in vitro → in cella approach, we identified functional allele-specific transcription factor motifs in the human PNMT promoter, including the human ortholog of a rat SP1/EGR1 motif (reviewed in Wong and Tank 2007) and a novel SOX17 motif.
G-367A motif and factors SP1 and EGR1
The G/C-rich SP1 motif is conserved across mammalian species and disrupted by common human variant G-367A (GCGGG[G/A]GGC, rs3764351; Fig. 2a), resulting in a loss of observed binding in EMSA experiments and a decrease in promoter enhancement in luciferase assays. An EGR1 motif is also likely in this region (Supplementary Fig. 1). We confirm regulatory effects of SP1 and EGR1 on this motif in both antibody disruption (Fig. 5) and ChIP (Fig. 6) experiments. The consistency of the effects across experiments, backed by extensive mammalian conservation of the motif, provides evidence supporting a functional polymorphism. Of note for the potential physiological significance of these findings, SP1 and EGR1 are highly expressed in PC12 chromaffin cells (GEO database GDS1038 and GDS1039 at http://www.ncbi.nlm.nih.gov/geo/).
G-161A motif and factor SOX17
At G-161A, a SOX17 motif conserved in primates (Fig. 2) is disrupted by the variant allele (TCC[G/A]GTGTG, rs876493), and the downregulatory response of the A allele at this motif was augmented by SOX17 + SP1 co-trans-activation. We documented an effect of SOX17 on the G-161 motif in both antibody disruption (Fig. 5) and ChIP (Fig. 6) experiments. SOX17 is also highly expressed in PC12 chromaffin cells (GEO GDS1039).
Perspectives for human trait determination
The significance of the results presented here is best understood in the context of the body of linkage and disease associations already identified for the PNMT gene. Indeed, not only is the PNMT locus within the genome-wide confidence interval for linkage with multiple stress traits in rodents (http://rgd.mcw.edu/), but PNMT promoter allelic associations occur for several human traits with likely derangements in adrenergic function, including the common diseases hypertension (Cui et al. 2003; Rana et al. 2007) and obesity (Peters et al. 2003). While published single-marker-on-trait associations for both promoter SNPs exist, the high degree of linkage disequilibrium (D′ > 0.9) between the SNPs makes it difficult to definitively ascribe a causal association to one of the two variants. Using the PC12 cell as a model of the human chromaffin cell, we were able to show allele-specific differences of promoter activity in the presence of transcription factors SP1, EGR1, and SOX17.
An earlier report on PNMT transcription using a longer (~ 1 kb) transfected human promoter (Ji et al. 2005) found similar rank-order activities of the two most common PNMT promoter haplotypes (G.G > A.A; p < 0.002), but no significant differences from G.G for the other haplotypes (p > 0.05) in PC12 (rat pheochromocytoma) and TE671 (human medulloblastoma) cells; however, that report did not pursue regulatory motifs at -367/-161. In a follow-up study, Ji et al. (2008) also explored the effect of a relatively rare polymorphism in the first PNMT intron (G280A) on plasma epinephrine as well as on PNMT transcription. Here we focused instead upon the PNMT proximal promoter, since the bulk of previous clinical associations implicated this region (Cui et al. 2003; Mann et al. 2001, 2002; Peters et al. 2003), the LD across locus is high, and common functional variation may influence large numbers of individuals.
References
Andersen MC, Engstrom PG, Lithwick S, Arenillas D, Eriksson P et al (2008) In silico detection of sequence variations modifying transcriptional regulation. PLoS Comput Biol 4:e5
Avery-Kiejda KA, Zhang XD, Adams LJ, Scott RJ, Vojtesek B et al (2008) Small molecular weight variants of p53 are expressed in human melanoma cells and are induced by the DNA-damaging agent cisplatin. Clin Cancer Res 14(6):1659–1668
Berezikov E, Guryev V, Cuppen E (2005) CONREAL web server: identification and visualization of conserved transcription factor binding sites. Nucleic Acids Res 33:W447–W450
Blanchette M, Kent WJ, Riemer C, Elnitski L, Smit AF et al (2004) Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14:708–715
Brudno M, Do CB, Cooper GM, Kim MF, Davydov E et al (2003) LAGAN and Multi-LAGAN: efficient tools for large-scale multiple alignment of genomic DNA. Genome Res 13:721–731
Chen Y, Rao F, Rodriguez-Flores JL, Mahapatra NR, Mahata M et al (2008) Common genetic variants in the chromogranin A promoter alter autonomic activity and blood pressure. Kidney Int 74:115–125
Cui J, Zhou X, Chazaro I, DeStefano AL, Manolis AJ et al (2003) Association of polymorphisms in the promoter region of the PNMT gene with essential hypertension in African Americans but not in whites. Am J Hypertens 16:859–863
Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30:207–210
Felsenstein J, Churchill GA (1996) A hidden Markov model approach to variation among sites in rate of evolution. Mol Biol Evol 13:93–104
Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL et al (2007) A second generation human haplotype map of over 3.1 million SNPs. Nature 449:851–861
Ji Y, Salavaggione OE, Wang L, Adjei AA, Eckloff B et al (2005) Human phenylethanolamine N-methyltransferase pharmacogenomics: gene re-sequencing and functional genomics. J Neurochem 95:1766–1776
Ji Y, Snyder EM, Fridley BL, Salavaggione OE, Moon I et al (2008) Human phenylethanolamine N-methyltransferase genetic polymorphisms and exercise-induced epinephrine release. Physiol Genomics 33:323–332
Kaneda N, Ichinose H, Kobayashi K, Oka K, Kishi F et al (1988) Molecular cloning of cDNA and chromosomal assignment of the gene for human phenylethanolamine N-methyltransferase, the enzyme for epinephrine biosynthesis. J Biol Chem 263:7672
Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH et al (2002) The human genome browser at UCSC. Genome Res 12:996–1006
Kepp K, Juhanson P, Kozich V, Ots M, Viigimaa M et al (2007) Resequencing PNMT in European hypertensive and normotensive individuals: no common susceptibility variants for hypertension and purifying selection on intron 1. BMC Med Genet 8:47
Kuhn RM, Karolchik D, Zweig AS, Wang T, Smith KE et al (2009) The UCSC Genome Browser Database: update 2009. Nucleic Acids Res 37:D755–D761
Liao WP, Uetzmann L, Burtscher I, Lickert H (2009) Generation of a mouse line expressing Sox17-driven Cre recombinase with specific activity in arteries. Genesis 47:476–483
Liu Y, Asakura M, Inoue H, Nakamura T, Sano M et al (2007) Sox17 is essential for the specification of cardiac mesoderm in embryonic stem cells. Proc Natl Acad Sci USA 104:3859–3864
Mann MB, Wu S, Rostamkhani M, Tourtellotte W, MacMurray J et al (2001) Phenylethanolamine N-methyltransferase (PNMT) gene and early-onset Alzheimer disease. Am J Med Genet 105:312–316
Mann MB, Wu S, Rostamkhani M, Tourtellotte W, MacMurray JP et al (2002) Association between the phenylethanolamine N-methyltransferase gene and multiple sclerosis. J Neuroimmunol 124:101–105
Matata BM, Galinanes M (2002) Peroxynitrite is an essential component of cytokines production mechanism in human monocytes through modulation of nuclear factor-kappa B DNA binding activity. J Biol Chem 277:2330–2335
Matsui T, Kanai-Azuma M, Hara K, Matoba S, Hiramatsu R et al (2006) Redundant roles of Sox17 and Sox18 in postnatal angiogenesis in mice. J Cell Sci 119:3513–3526
Oltra E, Pfeifer I, Werner R (2003) Ini, a small nuclear protein that enhances the response of the connexin43 gene to estrogen. Endocrinology 144:3148–3158
Park KS, Wells JM, Zorn AM, Wert SE, Whitsett JA (2006) Sox17 influences the differentiation of respiratory epithelial cells. Dev Biol 294:192–202
Peters WR, MacMurry JP, Walker J, Giese RJ Jr, Comings DE (2003) Phenylethanolamine N-methyltransferase G-148A genetic variant and weight loss in obese women. Obes Res 11:415–419
Rana BK, Insel PA, Payne SH, Abel K, Beutler E et al (2007) Population-based sample reveals gene-gender interactions in blood pressure in White Americans. Hypertension 49:96–106
Saito Y, Kanai Y, Sakamoto M, Saito H, Ishii H et al (2001) Expression of mRNA for DNA methyltransferases and methyl-CpG-binding proteins and DNA methylation status on CpG islands and pericentromeric satellite regions during human hepatocarcinogenesis. Hepatology 33:561–568
Sasaoka T, Kaneda N, Kurosawa Y, Fujita K, Nagatsu T (1989) Structure of human phenylethanolamine N-methyltransferase gene: existence of two types of mRNA with different transcription initiation sites. Neurochem Int 15:555–565
Thorisson GA, Smith AV, Krishnan L, Stein LD (2005) The International HapMap Project Web site. Genome Res 15:1592–1593
Vlieghe D, Sandelin A, De Bleser PJ, Vleminckx K, Wasserman WW et al (2006) A new generation of JASPAR, the open-access repository for transcription factor binding site profiles. Nucleic Acids Res 34:D95–D97
Wasserman WW, Sandelin A (2004) Applied bioinformatics for the identification of regulatory elements. Nat Rev Genet 5:276–287
Wingender E, Dietze P, Karas H, Knuppel R (1996) TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res 24:238–241
Wong DL, Tank AW (2007) Stress-induced catecholaminergic function: transcriptional and post-transcriptional control. Stress 10:121–130
Wu S, Comings DE (1999) Two single nucleotide polymorphisms in the promoter region of the human phenylethanolamine N-methyltransferase PNMT gene. Psychiatr Genet 9:187–188
Yamano E, Isowa T, Nakano Y, Matsuda F, Hashimoto-Tamaoki T et al (2008) Association study between reward dependence temperament and a polymorphism in the phenylethanolamine N-methyltransferase gene in a Japanese female population. Compr Psychiatry 49:503–507
Acknowledgments
Thanks to Manjula Mahata (UCSD Department of Medicine) for PC12 cells. This work was supported by the National Institutes of Health and the Department of Veterans Affairs. JLRF was funded by a NHGRI pre-doctoral fellowship. This work was supported by grants from the National Institutes of Health: HL58120, MD000220, and DK079337.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Supplementary Fig. 1
Putative polymorphic EGR1 binding motif flanking human PNMT promoter variant G-367A. For details of nomenclature and methods, see Fig. 1 caption (TIFF 1520 kb)
Rights and permissions
Open Access This is an open access article distributed under the terms of the Creative Commons Attribution Noncommercial License (https://creativecommons.org/licenses/by-nc/2.0), which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
About this article
Cite this article
Rodríguez-Flores, J.L., Zhang, K., Kang, S.W. et al. Conserved regulatory motifs at phenylethanolamine N-methyltransferase (PNMT) are disrupted by common functional genetic variation: an integrated computational/experimental approach. Mamm Genome 21, 195–204 (2010). https://doi.org/10.1007/s00335-010-9253-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-010-9253-y