Abstract
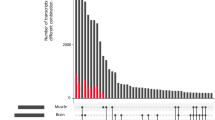
We established the Pig Genome Database (PiGenome) for pig genome research. The PiGenome integrates and analyzes all publicly available genome-wide data on pigs, including UniGenes, sequence tagged sites (STS) markers, quantitative trait loci (QTLs) data, and bacterial artificial chromosome (BAC) contigs. In addition, we produced 69,545 expressed sequence tags (ESTs) from the full-length enriched cDNA libraries of six tissues and 182 BAC contig sequences, which are also included in the database. QTLs, genetic markers, and BAC end-sequencing information were collected from public databases. The full-length enriched EST data were clustered and assembled into unique sequences, contigs, and singletons. The PiGenome provides functional annotation, identification of transcripts, mapping of coding sequences, and SNP information. It also provides an advanced search interface, a disease browser, alternative-splicing events, and a comparative gene map of the pig. A graphical map view and genome browser can map ESTs, contigs, BAC contigs (from the National Institute of Animal Science), Sino-Danish Pig Genome Project transcripts, and UniGene onto pig genome sequences which include our 182 BAC contigs and publically available BAC sequences of the Wellcome Trust Sanger Institute. The PiGenome is accessible at http://pigenome.nabc.go.kr/.


Similar content being viewed by others
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z et al (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Archibald AL, Haley CS, Brown JF, Couperwhite S, McQueen HA et al (1995) The PiGMaP consortium linkage map of the pig (Sus scrofa). Mamm Genome 6:157–175
Couronne O, Poliakov A, Bray N, Ishkhanov T, Ryaboy D et al (2003) Strategies and tools for whole-genome alignments. Genome Res 13:73–80
de Koning DJ, Janss LL, Rattink AP, van Oers PA, de Vries BJ et al (1999) Detection of quantitative trait loci for backfat thickness and intramuscular fat content in pigs (Sus scrofa). Genetics 152:1679–1690
Ewing B, Green P (1998) Base-calling of automated sequencer traces using phred. II. Error probabilities. Genome Res 8:186–194
Fang M, Hu X, Jiang T, Braunschweig M, Hu L et al (2005) The phylogeny of Chinese indigenous pig breeds inferred from microsatellite markers. Anim Genet 36:7–13
Florea L, Hartzell G, Zhang Z, Rubin GM, Miller W (1998) A computer program for aligning a cDNA sequence with a genomic DNA sequence. Genome Res 8:967–974
Gordon D, Abajian C, Green P (1998) Consed: a graphical tool for sequence finishing. Genome Res 8:195–202
Gorodkin J, Cirera S, Hedegaard J, Gilchrist MJ, Panitz F et al (2007) Porcine transcriptome analysis based on 97 non-normalized cDNA libraries and assembly of 1,021,891 expressed sequence tags. Genome Biol 8:R45
Grindflek E, Szyda J, Liu Z, Lien S (2001) Detection of quantitative trait loci for meat quality in a commercial slaughter pig cross. Mamm Genome 12:299–304
Hu J, Mungall C, Law A, Papworth R, Nelson JP et al (2001) The ARKdb: genome databases for farmed and other animals. Nucleic Acids Res 29:106–110
Hu ZL, Dracheva S, Jang W, Maglott D, Bastiaansen J et al (2005) A QTL resource and comparison tool for pigs: PigQTLDB. Mamm Genome 16:792–800
Huang X, Madan A (1999) CAP3: A DNA sequence assembly program. Genome Res 9:868–877
Jeon JT, Park EW, Jeon HJ, Kim TH, Lee KT et al (2003) A large-insert porcine library with sevenfold genome coverage: a tool for positional cloning of candidate genes for major quantitative traits. Mol Cells 16:113–116
Kim H, Lim D, Han BK, Sung S, Jeon M et al (2006a) ChickGCE: a novel germ cell EST database for studying the early developmental stage in chickens. Genomics 88:252–257
Kim TH, Kim NS, Lim D, Lee KT, Oh JH et al (2006b) Generation and analysis of large-scale expressed sequence tags (ESTs) from a full-length enriched cDNA library of porcine backfat tissue. BMC Genomics 7:36
Ovilo C, Perez-Enciso M, Barragan C, Clop A, Rodriquez C et al (2000) A QTL for intramuscular fat and backfat thickness is located on porcine chromosome 6. Mamm Genome 11:344–346
Ovilo C, Clop A, Noguera JL, Oliver MA, Barragan C et al (2002) Quantitative trait locus mapping for meat quality traits in an Iberian × Landrace F2 pig population. J Anim Sci 80:2801–2808
Rohrer GA, Alexander LJ, Hu Z, Smith TP, Keele JW et al (1996) A comprehensive map of the porcine genome. Genome Res 6:371–391
Rothschild MF (2003) From a sow’s ear to a silk purse: real progress in porcine genomics. Cytogenet Genome Res 102:95–99
Ruan J, Guo Y, Li H, Hu Y, Song F et al (2007) PigGIS: Pig Genomic Informatics System. Nucleic Acids Res 35:D654–D657
Stapleton M, Carlson J, Brokstein P, Yu C, Champe M et al (2002) A Drosophila full-length cDNA resource. Genome Biol 3:RESEARCH0080
Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S (1997) Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene 200:149–156
Szyda J, Liu Z, Grindflek E, Lien S (2002) Application of a mixed inheritance model to the detection of quantitative trait loci in swine. J Appl Genet 43:69–83
Uenishi H, Eguchi T, Suzuki K, Sawazaki T, Toki D et al (2004) PEDE (Pig EST Data Explorer): construction of a database for ESTs derived from porcine full-length cDNA libraries. Nucleic Acids Res 32:D484–D488
Venter JC, Adams MD, Sutton GG, Kerlavage AR, Smith HO et al (1998) Shotgun sequencing of the human genome. Science 280:1540–1542
Vodicka P, Smetana K Jr, Dvorankova B, Emerick T, Xu YZ et al (2005) The miniature pig as an animal model in biomedical research. Ann NY Acad Sci 1049:161–171
Wall DP, Fraser HB, Hirsh AE (2003) Detecting putative orthologs. Bioinformatics 19:1710–1711
Womack JE (2005) Advances in livestock genomics: opening the barn door. Genome Res 15:1699–1705
Zhuo D, Zhao WD, Wright FA, Yang HY, Wang JP et al (2001) Assembly, annotation, and integration of UNIGENE clusters into the human genome draft. Genome Res 11:904–918
Acknowledgments
This work was supported by a grant from the National Institute of Animal Science (H. Kim) and from the Biogreen21 Project (20050301034467) of the Korean Rural Development Administration (S.-K. Im).
Author information
Authors and Affiliations
Corresponding author
Additional information
The first three authors should be regarded as joint First Authors.
Rights and permissions
About this article
Cite this article
Lim, D., Cho, YM., Lee, KT. et al. The Pig Genome Database (PiGenome): an integrated database for pig genome research. Mamm Genome 20, 60–66 (2009). https://doi.org/10.1007/s00335-008-9156-3
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00335-008-9156-3