Abstract
Objectives
The accurate prediction of post-hepatectomy early recurrence in patients with hepatocellular carcinoma (HCC) is crucial for decision-making regarding postoperative adjuvant treatment and monitoring. We aimed to explore the feasibility of deep learning (DL) features derived from gadoxetate disodium (Gd-EOB-DTPA) MRI, qualitative features, and clinical variables for predicting early recurrence.
Methods
In this bicentric study, 285 patients with HCC who underwent Gd-EOB-DTPA MRI before resection were divided into training (n = 195) and validation (n = 90) sets. DL features were extracted from contrast-enhanced MRI images using VGGNet-19. Three feature selection methods and five classification methods were combined for DL signature construction. Subsequently, an mp-MR DL signature fused with multiphase DL signatures of contrast-enhanced images was constructed. Univariate and multivariate logistic regression analyses were used to identify early recurrence risk factors including mp-MR DL signature, microvascular invasion (MVI), and tumor number. A DL nomogram was built by incorporating deep features and significant clinical variables to achieve early recurrence prediction.
Results
MVI (p = 0.039), tumor number (p = 0.001), and mp-MR DL signature (p < 0.001) were independent risk factors for early recurrence. The DL nomogram outperformed the clinical nomogram in the training set (AUC: 0.949 vs. 0.751; p < 0.001) and validation set (AUC: 0.909 vs. 0.715; p = 0.002). Excellent DL nomogram calibration was achieved in both training and validation sets. Decision curve analysis confirmed the clinical usefulness of DL nomogram.
Conclusion
The proposed DL nomogram was superior to the clinical nomogram in predicting early recurrence for HCC patients after hepatectomy.
Key Points
• Deep learning signature based on Gd-EOB-DTPA MRI was the predominant independent predictor of early recurrence for hepatocellular carcinoma (HCC) after hepatectomy.
• Deep learning nomogram based on clinical factors and Gd-EOB-DTPA MRI features is promising for predicting early recurrence of HCC.
• Deep learning nomogram outperformed the conventional clinical nomogram in predicting early recurrence.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Hepatic resection is the first-line treatment for patients with early-stage HCC and well-preserved liver function [1, 2]. However, HCC recurrence rate reaches 70% 5 years after surgery [3]. More than 80% of recurrences are intrahepatic, including intrahepatic metastases from a primary tumor (considered true recurrence) and de novo multicentric metastasis [4]. The poor prognosis of patients is related to intrahepatic metastases and mainly presents as early recurrence (within 2 years), whereas late recurrence (> 2 years) is more likely associated with underlying liver diseases, such as cirrhosis [5, 6]. Thus, early recurrence risk assessment in patients with HCC is clinically relevant.
Numerous tumor factors have been identified as predictors of early recurrence, such as microvascular invasion (MVI), surgical margin, tumor size, high lobular hepatitis activity, and poor Edmondson-Steiner grade [7,8,9,10]. However, most of the risk factors can only be obtained by postoperative pathology, and thus cannot be used to assess prognosis and develop treatment plans before hepatectomy. Medical imaging is a routine preoperative examination for patients with HCC. Previous studies have shown that quantitative parameters measured from medical imaging equipment, such as metabolic parameters, apparent diffusion coefficient values, and liver stiffness values from positron emission tomography/computed tomography [11], diffusion-weighted imaging [12], and magnetic resonance elastography [13], respectively, showed great efficiency and high clinical practicability in predicting the early recurrence of HCC after hepatectomy. Although magnetic resonance elastography is accurate in detecting liver fibrosis, the widespread application of this imaging modality in patients with HCC is limited due to its low specificity and its dependence on quantitative parameters; however, the cutoff of parameters was lack of criterion. Gadoxetate disodium (Gd-EOB-DTPA), a hepatobiliary-specific contrast agent that is widely used in patients with HCC, can better capture the perfusion and functional alterations, and hence may be more sensitive and accurate in HCC detection. Previous studies [14, 15] have also revealed that Gd-EOB-DTPA MRI is better than multidetector computed tomography (CT) and MRI using other contrast agents.
Several imaging features observed on Gd-EOB-DTPA MRI, including rim enhancement, arterial peritumoral enhancement, non-smooth tumor margin, satellite nodule, and peritumoral hypointensity on hepatobiliary phase (HBP) images [16, 17], are associated with early recurrence in patients with HCC; nevertheless, early recurrence prediction using MR features may be subjective and dependent on radiologist experience. Qualitative MR features are limited by image grayscale recognition, and therefore, much information related to tumor heterogeneity is lost.
Radiomics, involving the high-throughput extraction and mining of quantitative imaging features, is thought to capture the histological heterogeneity inherent to solid tumors [18]. In contrast to invasive tissue biomarkers, quantitative features retrieved from CT or MR images have demonstrated improved diagnostic and prognostic precision in patients with HCC, such as the preoperative prediction of MVI and recurrence-free survival [19]. However, more recent studies have shifted towards the field of deep learning (DL).
DL, a subset of machine learning, is a new diagnostic technology for mining internal information from medical images. DL can be applied to tumor segmentation [20], prognosis prediction [21, 22], and treatment response evaluation [23] by automatically extracting deep-learned or high-order image features. Among them, convolutional neural network (CNN) is famous for handling image classification tasks [24]; the three major operations of CNNs are convolution, activation, and pooling, and the entire process can be divided into two steps: the forward computation and the back propagation [25]. Additionally, DL has provided a preoperative prediction tool to guide postoperative clinical decision‐making regarding patients with HCC, such as HCC recurrence prediction after liver transplantation [26] and prognostic factor exploration in HCC pathological images [27]. Thus, the direct use of DL-based image features to predict prognosis would provide a promising non-invasive method to better individualize patient treatment.
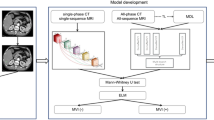
Therefore, we aimed to investigate the feasibility of deep features extracted from Gd-EOB-DTPA MR images for predicting the early recurrence of HCC after curative resection. Furthermore, we evaluated the predictive performance of the DL-based nomogram incorporating deep features and significant clinical factors.
Materials and methods
Patients and dataset
Ethical approval was obtained for this retrospective study, and the requirement for informed consent was waived. Patients with suspected HCC who underwent Gd-EOB-DTPA MRI between January 2012 and September 2018 prior to curative resection were consecutively included. This study was performed at two centers: Sun Yat-Sen University Cancer Center (center 1) and Southern Medical University affiliated Zhujiang Hospital (center 2). The inclusion criteria were as follows: patients (a) with pathological confirmation of HCC; (b) with Barcelona Clinic Liver Cancer stage 0, A, or B HCC; (c) who received no previous anti-cancer treatment; and (d) who underwent Gd-EOB-DTPA MRI of the liver within 1 month before surgery. The exclusion criteria were as follows: patients (a) with recurrent HCC or combined with hepatocyte cholangiocarcinoma or metastatic tumor in the liver; (b) with radiographic macrovascular invasion or extrahepatic metastasis; (c) with incomplete clinical, radiological, pathological, or follow-up data; and (d) who died due to postoperative complications or liver cancer rupture within 2 weeks (Fig. 1). All patients including 227 patients in center 1 and 58 patients in center 2 were randomly divided into training and validation sets at in a 7:3 ratio.
Baseline clinicopathological data were collected from electronic medical records. Clinical data included demographics, time to early recurrence, and Barcelona Clinic Liver Cancer stage. Laboratory features included neutrophil count as well as hepatitis B virus DNA, α-fetoprotein (AFP), alanine aminotransferase, aspartate aminotransferase (AST), and γ-glutamyl transpeptidase levels. Pathologic data included MVI, defined as tumor emboli in a vascular space lined by endothelial cells on microscopy [28], tumor number, and histologic grade, which was evaluated as well differentiated, moderately differentiated, or poorly differentiated.
Follow-up surveillance and clinical endpoint
All patients were followed up for at least 2 years after curative resection. Patients were screened for tumor recurrence through serum AFP level evaluation, ultrasonography, contrast-enhanced CT, or MRI of the chest and abdomen in the first month after surgery, once every 3 months thereafter during the first year, and every 6 months thereafter. The censored follow-up date was October 1, 2020.
The study endpoint was early recurrence, which was defined as one or more of the following events occurring within 2 years after curative resection: (a) presence of new hepatic lesions with typical imaging findings of HCC; (b) atypical imaging findings with biopsy or re-postoperative pathology-confirmed HCC, or postoperative transarterial chemoembolization (TACE) revealed tumor staining; and (c) extrahepatic metastases confirmed by typical imaging features or histological analysis.
MRI acquisition and preprocessing
MR images of the arterial phase (AP), portal venous phase (PVP), and HBP were collected in this study. The MRI machines and parameters provided in sequences and parameters are listed in Supplementary Methods and Supplementary Table 1. Before the quantitative analysis, we preprocessed the MR images considering the influence of different MRI scanning parameters, protocols, and individual differences. First, an offset field correction by N4 algorithm in 3D Slicer software was applied to correct the inhomogeneity in gray level. Then, we mapped the gray values to a range of 0–255, to reduce the influence of different gray scales on gray quantization.
Qualitative analysis of MR images
The qualitative analysis of MR image features was independently performed by three abdominal radiologists (M.Y., B.Z., and Z.J.G. with 5, 10, and 16 years of abdominal diagnosis experience, respectively). The radiologists were blinded to the radiological and pathologic reports. Reader 1 (M.Y.) observed the features twice, and the second observation was performed 2 weeks after the first observation.
The MR features included the following: (a) tumor size, defined as the maximum diameter on transverse HBP images; (b) AP enhancement type (types 1 [homogeneous enhancement pattern with no increased arterial blood flow], 2 [homogeneous enhancement with increased arterial blood flow], 3 [heterogeneous enhancement containing non-enhanced areas], 4 [heterogeneous enhancement pattern with irregular ring-like structures] [29,30,31], and 5 [heterogeneous and hypointense enhancement pattern]); (c) capsule appearance (peripheral rim of uniform and smooth hyperenhancement in the portal or delayed phase, which is categorized into three groups [absent, incomplete, and complete]) [32]; (d) hypodense halo (a rim of hypointensity partially or wholly surrounding the tumor); (e) intratumor necrosis (a low signal on T1-weighted imaging, a high signal on T2-weighted imaging, and a low signal on all enhanced phases); (f) satellite nodules, defined as small (< 2 cm) tumor nodules close (< 2 cm) to the main tumor [33]; (g) peritumoral hypointensity, defined as flame-like or wedge-shaped hypointense areas of the hepatic parenchyma around the tumor on HBP images [34]. Figure 2 shows the MR features.
Definitions of features and representative MR images. Abbreviations: AP, arterial phase; Type 1, a homogeneous enhancement pattern with no increased arterial blood flow; Type 2, a homogeneous enhancement with increased arterial blood flow; Type 3, a heterogeneous enhancement included non-enhanced areas; Type 4, a heterogeneous enhancement pattern with irregular ring-like structures; Type 5, a heterogeneous and hypointense enhancement pattern; HBP, hepatobiliary phase
Image segmentation and DL feature extraction
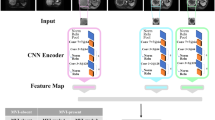
The regions of interest were delineated around the tumor boundary on the largest dimension. A state-of-the-art architecture VGGNet-19 was then applied to extract 1472 DL features from the AP, PVP, and HBP images. The DL network contains five convolutional layers, four max-pooling layers, three fully connected layers, and a softmax layer. The DL workflow is shown in Fig. 3, and more details are provided in Supplementary Methods.
Feature selection and DL signature development
To select the features strongly related to early recurrence, the DL features were subjected to the following steps: feature value preconditioning, de-redundancy, and dimensionality reduction; thereafter, machine learning methods were used to predict the status of outcome events and establish a DL signature that can predict early recurrence. All features were first normalized to the range of [0,1] by the minimum–maximum normalization method. Moreover, Spearman correlation analysis was added to retain DL features associated with the early recurrence of HCC (p < 0.05). Subsequently, the Pearson correlation coefficient (r) was used to remove one redundant feature with a lower r from the feature pairs (r > 0.9). The highly predictive features obtained were further screened by variance analysis, recursive feature elimination (RFE), and Relief algorithm. Five types of classifiers, namely random forest (RF), support vector machine, least absolute shrinkage and selection operator logistic regression (LASSO), AdaBoost, and Gaussian process (GP), were compared to identify the outcome status of early recurrence for every phase of the DL features.
Clinical and DL analysis
Univariate logistic regression analysis was performed in the training set, and significant variables (p < 0.05) were entered into the multivariate logistic regression using the forward likelihood ratio method to identify the independent risk factors for early recurrence. A two-sided p < 0.05 was considered statistically significant. The nomogram was plotted based on multivariate logistic regression analysis findings.
Collinearity analysis of conventional clinical factors and DL signatures was also performed. The evaluation factors were tolerance and variance inflation factor (VIF); a tolerance value < 0.1 or a VIF value > 5 was considered to indicate collinearity between two variables.
We calculated the radiomics quality score (https://www.radiomics.world/rqs2) to assess the methodology, analysis, and reporting of our DL study [35].
Statistical analysis
Comparisons between the training and validation sets were conducted using the chi-square test or Fisher’s exact test for categorical variables, whereas the Mann–Whitney U test was used for continuous variables. Intra-reader agreements of the qualitative MR imaging features were assessed using Cohen’s κ coefficient, and the inter-reader agreement was assessed by Fleiss’ κ statistics across three readers. Kappa (κ) statistics were qualitatively stratified by κ = 0.00–0.20, poor agreement; κ = 0.21–0.40, fair agreement; κ = 0.41–0.60, moderate agreement; κ = 0.61–0.80, good agreement; and κ = 0.81–1.00, excellent agreement [35].
The receiver operating characteristic curve analysis was employed to calculate the area under the curve (AUC), accuracy, sensitivity, and specificity. Comparisons between different DL signatures and models were implemented using the DeLong test. Model fit was assessed via calibration plots using 1000 bootstrap resamples. The clinical utility of the models was evaluated using decision curve analysis. Software and packages for statistical analyses are provided in Supplementary Methods. All statistical tests were two-sided, and a p < 0.05 was considered statistically significant.
Results
Clinical characteristics
We included 285 patients (men, n = 254; median age, 54.0 years; range, 13–79 years) whose data were divided into the training set (n = 195) and validation set (n = 90). Early recurrence occurred in 77 (27.0%) patients, and there was no difference in the early recurrence rate between the training set (27.7%, 54/195) and validation set (25.6%, 23/90). A total of 364 tumors were found in all patients, including 1, 2, 3, and 4 tumors in 223, 47, 13, and 2 patients, respectively. The clinical characteristics of the two groups are shown in Supplementary Table 2. No statistically significant difference was observed in baseline clinicopathological variables, pathologic variables, and qualitative MR features between the two sets (p = 0.113–1.000) except for the factor of etiology (p = 0.012).
Among all HCC patients in the early recurrence group, there were 52 (67.5%) patients with intrahepatic recurrence, including 48 (62.3%), 3 (3.9%), and 1 (1.3%) patients with typical CT/MR finding, re-postoperatively confirmed HCC, and TACE-revealed tumor staining, respectively; and 30 (39%) patients with extrahepatic recurrence, including 25 (32.5%) and 4 (5.2%) with typical CT/MR finding and histological analysis, respectively. Additionally, 5 (6.5%) patients had concurrent intra- and extrahepatic recurrences. The median time to early recurrence was 8 months (range, 1–24 months). The recurrence details are provided in Supplementary Table 6.
Intra- and inter-reader agreement for qualitative MR images
Supplementary Table 5 presents the percentages of all features identified by the three readers as well as their intra- and inter-reader agreements and κ statistics for each imaging. Each MR feature showed an almost perfect intra-reader agreement (κ = 0.81–1.00). Regarding the inter-reader agreement among the three readers, the hypodense halo, peritumoral hypointensity, satellite nodules, capsule appearance, and intratumor necrosis showed good agreement (κ = 0.72–0.79); AP enhancement type and tumor size showed excellent agreement (κ = 0.84 and 0.97, respectively).
DL signature development and validation
For identifying the outcome status of early recurrence in every phase of AP, PVP, and HBP features, the performance comparisons among five classifiers are shown in Fig. 4. It manifested that the GP classifier achieved the best performance for all the sequences with the AUCs of 0.826 (AP), 0.854 (PVP), and 0.888 (HBP), while the AdaBoost classifier showed almost no discrimination ability with the AUCs of 0.519 (AP), 0.503 (PVP), and 0.485 (HBP). In total, 99 DL features were selected from AP images using RFE, 93 DL features were screened from PVP images using Relief, and 99 DL features were identified from HBP images using Relief. All the optimal single-layered DL signatures were constructed using GP classifiers. All DL signatures showed significant differences between the two groups (all p < 0.05) in the training set (Table 1). Collinearity analysis was performed for variables with p < 0.05 after univariate analysis. The tolerance range of each variable was 0.750–0.970, and the VIF range was 1.031–1.333, indicating no collinearity between the variables (Supplementary Table 3).
Performance comparisons among the five classifiers for the validation set: A AP images; B PVP images; C HBP images. Abbreviations: AP, arterial phase; PVP, portal venous phase; HBP, hepatobiliary phase; GP, Gaussian process; LASSO, least absolute shrinkage and selection operator logistic regression; RF, random forest; SVM, support vector machine
The AP DL signature achieved an AUC of 0.882 (95% confidence interval [CI]: 0.823–0.941) in the training set and 0.826 (95% CI: 0.755–0.897) in the validation set (Table 2); the PVP DL signature yielded an AUC of 0.822 (95% CI: 0.753–0.891) in the training set and 0.854 (95% CI: 0.780–0.928) in the validation set (Table 2), whereas the HBP-DL signature demonstrated an AUC of 0.909 (95% CI: 0.860–0.958) in the training set and 0.888 (95% CI: 0.833–0.943) in the validation set (Table 2). The multi-sequence MR (mp-MR) DL signature performance improved in both training and validation sets, with AUCs of 0.929 (95% CI: 0.829–0.966) and 0.894 (95% CI: 0.811–0.977) in the training and validation sets, respectively (Table 2). The DeLong test demonstrated that the mp-MR DL signature performance was better than that of the AP DL signature in both training and validation sets (p < 0.05) (Table 2).
Establishment of clinical and DL nomograms
After univariate analysis, six clinical factors, namely neutrophil count, AST level, γ-glutamyl transpeptidase level, MVI, tumor number, and histologic grade (p < 0.05), were candidate factors for multivariate analysis (Supplementary Table 4); however, only neutrophil count, AST level, and MVI were identified as independent risk factors for early recurrence of HCC. The clinical nomogram was constructed based on the three factors (Table 3; Fig. 5A) which achieved an AUC of 0.751 (95% CI: 0.674–0.827) in the training set and 0.715 (95% CI: 0.586–0.843) in the validation set (Table 4; Fig. 6A, B).
Clinical and deep learning nomograms. Clinical nomogram (A) was developed with NE, AST level, and MVI. Deep learning nomogram (B) was developed with MVI, tumor number, and mp-MR DL signature. Abbreviations: NE, neutrophil count. AST, aspartate aminotransferase. MVI, microvascular invasion; mp-MR, multiple sequences magnetic resonance; DL, deep learning
Discrimination, calibration, and clinical usefulness of the nomograms. The receiver operating characteristic curves for clinical nomogram and deep learning nomogram in the training (A) and validation (B) sets. Calibration curves for the clinical and deep learning nomograms in the training (C) and validation (D) sets. The y-axis represents the actual early recurrence rate, the x-axis represents the predicted early recurrence, and the diagonal dashed line represents the ideal prediction by a perfect model. Decision curve analysis for the clinical and deep learning nomograms in the training (E) and validation (F) sets. The y-axis represents the net benefits, and the x-axis represents the threshold probability. The deep learning nomogram (red line) had a good net benefit compared with the clinical nomogram (blue line), simple strategies such as follow-up of all patients (gray line), or no patients (horizontal black line) across the majority range of threshold probabilities. Abbreviation: DL, deep learning
Thereafter, the above-mentioned five clinical factors and the mp-MR DL signature were included in the multivariate analysis, and tumor number, MVI, and mp-MR DL signature were identified as independent risk factors for early recurrence (p < 0.05). We developed a DL nomogram incorporating the tumor number, MVI, and mp-MR DL signature (Table 3; Fig. 5B), which significantly outperformed the clinical nomogram, yielding an AUC of 0.949 (95% CI: 0.919–0.980) in the training set and 0.909 (95% CI: 0.842–0.976) in the validation set (Table 4; Fig. 6A, B). The DeLong test showed a significant difference between the clinical and DL nomograms in the training set (p < 0.001) and validation set (p = 0.002) (Table 4). Figure 6C and D demonstrate good DL nomogram calibration. The decision curve showed that the DL nomogram had a higher net benefit than the clinical nomogram (Fig. 6E, F). This DL study scored 40 points (60.6%) (Supplementary Material_RQS).
Discussion
We used the DL approach to explore the informative features from Gd-EOB-DTPA MRI images that were associated with early recurrence of HCC and established three single-layered DL signatures and an mp-MR DL signature fused with three-phase MR sequences. The results showed that the mp-MR DL signature was better than the three single-layered DL signatures. Subsequently, the DL nomogram was constructed by integrating tumor number, MVI, and the mp-MR DL signature, which achieved higher predictive accuracy and better net benefit than the clinical nomogram. This study demonstrated the incremental value of the DL nomogram compared with the conventional clinical nomogram.
Previous studies also revealed that Gd-EOB-DTPA MRI had a significantly higher sensitivity and overall accuracy for HCC diagnosis, especially small lesions, than multiphasic CT without substantial loss of specificity [36, 37]. Hence, we chose Gd-EOB-DTPA MRI instead of CT images to predict the early recurrence in patients with HCC. As recent studies reported [38, 39], obtaining high stability of features from different MR scanners becomes an increasingly common challenge in radiomics or DL field. As the data in our study were obtained from different MR scanners, we did offset field correction and gray normalization to reduce the potential affect. The preprocessing procedure is usually conducted in other studies [10, 40, 41]. The challenge of eliminating feature variability may be addressed through optimization tools developed for DL, via standardization of imaging protocols [42], and through prospective studies. In addition, we demonstrated that the HBP signature yielded a higher AUC and sensitivity than AP signature and PVP signature. Well-defined tumor margins on HBP images allow for a more accurate tumor delineation than those on AP and PVP images, in which the tumor margins could be affected by peritumoral enhancement, capsule appearance, and a hypodense halo.
Some previous studies have explored the feasibility of predicting early postoperative HCC recurrence. An et al. [17] and Zhang et al. [44] developed nomograms containing clinic-radiological variables to predict the early recurrence in patients with HCC, achieving AUCs of 0.783–0.846 in the validation cohorts. Chan et al. [45] constructed a preoperative model (early recurrence after surgery for liver tumor [ERASL]-pre) and a postoperative model (ERASL-post) based on gender, AFP level, MVI, tumor size, and tumor number, with AUCs ranging from 0.614 to 0.736 for the ERASL-pre and 0.653 to 0.763 for the ERASL-post in the derivation cohorts from different countries or districts. In our study, we also built a clinical nomogram and the diagnostic efficacy was similar to those in previous studies, with AUCs of 0.751 and 0.715 in the training and validation sets, respectively. Although these data are commonly and easily available in clinical practice, the two major problems encountered were non-uniformity in the standardization of clinical factors and the inability to handle high-dimension features using traditional statistical methods [46].
In recent years, artificial intelligence has emerged as an effective tool to demonstrate multi-modal patient data [47]. Radiomics is a recently emerged technology that extracts a large number of quantitative image features from standard-of-care medical imaging using data-characterization algorithms. Zhang et al. [48] and Zhao et al. [40] constructed a nomogram by integrating radiomic score and clinic-radiological factors, with AUCs of 0.844 and 0.873, respectively. Kim et al. [49] developed a combined clinicopathologic-radiomic model via random survival forest algorithm, which acquired a C-index of 0.716. In the latest research, the DL methods have outperformed the radiomics features in many tasks including lesion detection [50], prognosis prediction [51], and multimodal image registration [52]. The image-based DL technology has been widely applied in the HCC field of mass differentiation, treatment response, and prognosis [10, 53,54,55]. Song et al. [56] established a DL model using CNN in eight MRI sequences to predict the presence of MVI, which acquired an AUC of 0.931. Zhang et al. [57] constructed an integrated nomogram combining clinical features and DL signatures based on contrast CT to improve overall survival prediction in HCC patients treated with TACE plus sorafenib, with a C-index of 0.730 in the validation set. Based on the successful applications of DL in patients with HCC, we explored the VGGnet-19, which generally performs a more robust and automatic image analysis without export’s intervention [58], to extend the associations between DL and prognosis of HCC. Additionally, we employed 2D regions of interest to establish the DL signatures, which showed great performance; this finding corroborates with previous studies [57, 59, 60], wherein AUCs of 0.826–0.894 were achieved in the validation set. It is probably because the DL signatures could make predictions by capturing both global and local features of tumors, and it comprehensively reflected on the tumor size and heterogeneity, which were established prognostic factors [61, 62]. Thus, it may also explain the lack of tumor size as an independent risk factor because the DL signatures have already contained the information of tumor size which belongs to the tumor global feature. Nevertheless, this is subject to our future research.
In the field of feature engineering, different machine learning–based dimensionality reduction techniques have distinct mathematical senses and inherent limitations; thus, multiple algorithms should be combined to select robust features [42, 63]. Previous studies also proved that the combination of different dimensional reduction methods with several machine learning methods could maximize model diagnostic performance. Dai et al. [64] reported that feature selection and modeling methods could potentially affect prediction models. The optimal radiomic model for MVI evaluation was constructed using a gradient boosting decision tree classifier, which outperformed logistic regression, support vector machine, and random forest. Using 21 combination methods (including three feature selection methods and seven classification methods), Ni et al. [65] identified LASSO plus gradient boosting decision tree as the optimal combination for predicting MVI in patient with HCC. In the present study, we compared three feature selection methods with five classification methods to determine the best combination and found that RFE or Relief combined with GP classifier achieved the optimal performance in building DL signatures. Consequently, it is necessary to implement performance comparisons of different machine learning methods, which was absent in previous deep learning studies for HCC [43, 56, 57].
Our study has several limitations. First, the retrospective nature of the study may induce inevitable selection bias. Second, although we collected patient data from two centers, we were unable to perform an external validation because the sample size of center 2 was small; thus, multicenter studies are needed to validate the generalization ability of the proposed DL nomogram. Third, our study did not explore the role of DL features extracted from non-contrast MR sequences such as T1-weighted imaging, T2-weighted imaging, and diffusion-weighted imaging for predicting prognosis. Finally, the value of the DL model for improving long-term survival in patients with HCC remains unclear, and the differences between DL model–assisted and non-assisted practices warrant further study to prove the clinical applicability of the DL model.
In conclusion, we developed a DL nomogram that incorporates clinical factors and Gd-EOB-DTPA MRI biomarkers; furthermore, the DL nomogram was more effective in early recurrence prediction and postoperative surveillance than the traditional clinical nomogram in patients with HCC following surgical resection.
Data Availability
The data that support the findings of this study are available on reasonable request from the corresponding author. The data are not publicly available due to privacy or ethical restrictions.
Abbreviations
- AFP:
-
α-fetoprotein
- AP:
-
Arterial phase
- AST:
-
Aspartate aminotransferase
- AUC:
-
Area under the curve
- CNN:
-
Convolutional neural network
- DL:
-
Deep learning
- ERASL:
-
Early recurrence after surgery for liver tumor
- Gd-EOB-DTPA:
-
Gadoxetate disodium
- GP:
-
Gaussian process
- HBP:
-
Hepatobiliary phase
- HCC:
-
Hepatocellular carcinoma
- LASSO:
-
Least absolute shrinkage and selection operator logistic regression
- mp-MR:
-
Multiple sequences magnetic resonance
- MVI:
-
Microvascular invasion
- PVP:
-
Portal venous phase
- RFE:
-
Recursive feature elimination
- TACE:
-
Transarterial chemoembolization
References
Marasco G, Colecchia A, Colli A et al (2019) Role of liver and spleen stiffness in predicting the recurrence of hepatocellular carcinoma after resection. J Hepatol 70:440–448
Bruix J, Sherman M (2011) Management of hepatocellular carcinoma: an update. Hepatology 53:1020–1022
Ishizawa T, Hasegawa K, Aoki T et al (2008) Neither multiple tumors nor portal hypertension are surgical contraindications for hepatocellular carcinoma. Gastroenterology 134:1908–1916
Tung-Ping Poon R, Fan ST, Wong J (2000) Risk factors, prevention, and management of postoperative recurrence after resection of hepatocellular carcinoma. Ann Surg 232:10–24
Portolani N, Coniglio A, Ghidoni S et al (2006) Early and late recurrence after liver resection for hepatocellular carcinoma: prognostic and therapeutic implications. Ann Surg 243:229–235
Poon RT, Fan ST, Ng IO, Lo CM, Liu CL, Wong J (2000) Different risk factors and prognosis for early and late intrahepatic recurrence after resection of hepatocellular carcinoma. Cancer 89:500–507
Lee S, Kang TW, Song KD et al (2021) Effect of microvascular invasion risk on early recurrence of hepatocellular carcinoma after surgery and radiofrequency ablation. Ann Surg 273:564–571
Hirokawa F, Hayashi M, Asakuma M, Shimizu T, Inoue Y, Uchiyama K (2016) Risk factors and patterns of early recurrence after curative hepatectomy for hepatocellular carcinoma. Surg Oncol 25:24–29
Kwon SK, Yun SS, Kim HJ, Lee DS (2014) The risk factors of early recurrence after hepatectomy in hepatocellular carcinoma. Ann Surg Treat Res 86:283–288
Li X, Qi Z, Du H et al (2021) Deep convolutional neural network for preoperative prediction of microvascular invasion and clinical outcomes in patients with HCCs. Eur Radiol. https://doi.org/10.1007/s00330-021-08198-w
Miao W, Nie P, Yang G et al (2021) An FDG PET/CT metabolic parameter-based nomogram for predicting the early recurrence of hepatocellular carcinoma after liver transplantation. Eur J Nucl Med Mol Imaging 48:3656–3665
Choi SY, Kim SH, Park CK et al (2018) Imaging features of gadoxetic acid-enhanced and diffusion-weighted MR imaging for identifying cytokeratin 19-positive hepatocellular carcinoma: a retrospective observational study. Radiology 286:897–908
Cho HJ, Kim B, Kim HJ et al (2020) Liver stiffness measured by MR elastography is a predictor of early HCC recurrence after treatment. Eur Radiol 30:4182–4192
Kim BR, Lee JM, Lee DH et al (2017) Diagnostic performance of gadoxetic acid-enhanced liver MR imaging versus multidetector CT in the detection of dysplastic nodules and early hepatocellular carcinoma. Radiology 285:134–146
Lee YJ, Lee JM, Lee JS et al (2015) Hepatocellular carcinoma: diagnostic performance of multidetector CT and MR imaging-a systematic review and meta-analysis. Radiology 275:97–109
Lee S, Kim SH, Lee JE, Sinn DH, Park CK (2017) Preoperative gadoxetic acid-enhanced MRI for predicting microvascular invasion in patients with single hepatocellular carcinoma. J Hepatol 67:526–534
An C, Kim DW, Park YN, Chung YE, Rhee H, Kim MJ (2015) Single hepatocellular carcinoma: preoperative MR imaging to predict early recurrence after curative resection. Radiology 276:433–443
Lambin P, Rios-Velazquez E, Leijenaar R et al (2012) Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer 48:441–446
Chong HH, Yang L, Sheng RF et al (2021) Multi-scale and multi-parametric radiomics of gadoxetate disodium-enhanced MRI predicts microvascular invasion and outcome in patients with solitary hepatocellular carcinoma ≤ 5 cm. Eur Radiol 31:4824–4838
Ben Naceur M, Akil M, Saouli R, Kachouri R (2020) Fully automatic brain tumor segmentation with deep learning-based selective attention using overlapping patches and multi-class weighted cross-entropy. Med Image Anal 63:101692
Zhang W, Fang M, Dong D et al (2020) Development and validation of a CT-based radiomic nomogram for preoperative prediction of early recurrence in advanced gastric cancer. Radiother Oncol 145:13–20
Lin T, Mai J, Yan M, Li Z, Quan X, Chen X (2021) A nomogram based on CT deep learning signature: a potential tool for the prediction of overall survival in resected non-small cell lung cancer patients. Cancer Manag Res 13:2897–2906
Liu D, Liu F, Xie X et al (2020) Accurate prediction of responses to transarterial chemoembolization for patients with hepatocellular carcinoma by using artificial intelligence in contrast-enhanced ultrasound. Eur Radiol 30:2365–2376
Murphy A, Skalski M, Gaillard F (2018) The utilisation of convolutional neural networks in detecting pulmonary nodules: a review. Br J Radiol 91:20180028
Shin HC, Roth HR, Gao M et al (2016) Deep convolutional neural networks for computer-aided detection: CNN architectures, dataset characteristics and transfer learning. IEEE Trans Med Imaging 35:1285–1298
Nam JY, Lee JH, Bae J et al (2020) Novel model to predict HCC recurrence after liver transplantation obtained using deep learning: a multicenter study. Cancers (Basel) 12:2791
Shi JY, Wang X, Ding GY et al (2021) Exploring prognostic indicators in the pathological images of hepatocellular carcinoma based on deep learning. Gut 70:951–961
Roayaie S, Blume IN, Thung SN et al (2009) A system of classifying microvascular invasion to predict outcome after resection in patients with hepatocellular carcinoma. Gastroenterology 137:850–855
Kawamura Y, Ikeda K, Seko Y et al (2011) Heterogeneous type 4 enhancement of hepatocellular carcinoma on dynamic CT is associated with tumor recurrence after radiofrequency ablation. AJR Am J Roentgenol 197:W665-673
Kawamura Y, Ikeda K, Hirakawa M et al (2010) New classification of dynamic computed tomography images predictive of malignant characteristics of hepatocellular carcinoma. Hepatol Res 40:1006–1014
Shimizu R, Tamai H, Mori Y et al (2016) The arterial tumor enhancement pattern on contrast-enhanced computed tomography is associated with primary cancer death after radiofrequency ablation for small hepatocellular carcinoma. Hepatol Int 10:328–339
Huang M, Liao B, Xu P et al (2018) Prediction of microvascular invasion in hepatocellular carcinoma: preoperative Gd-EOB-DTPA-dynamic enhanced MRI and histopathological correlation. Contrast Media Mol Imaging 2018:9674565
Ji G-W, Zhu F-P, Xu Q et al (2020) Radiomic features at contrast-enhanced CT predict recurrence in early stage hepatocellular carcinoma: a multi-institutional study. Radiology 294:568–579
Kim KA, Kim MJ, Jeon HM et al (2012) Prediction of microvascular invasion of hepatocellular carcinoma: usefulness of peritumoral hypointensity seen on gadoxetate disodium-enhanced hepatobiliary phase images. J Magn Reson Imaging 35:629–634
Li W, Li L, Zhuang BW et al (2021) Inter-reader agreement of CEUS LI-RADS among radiologists with different levels of experience. Eur Radiol 31:6758–6767
Liu X, Jiang H, Chen J, Zhou Y, Huang Z, Song B (2017) Gadoxetic acid disodium-enhanced magnetic resonance imaging outperformed multidetector computed tomography in diagnosing small hepatocellular carcinoma: a meta-analysis. Liver Transpl 23:1505–1518
Onishi H, Kim T, Imai Y et al (2012) Hypervascular hepatocellular carcinomas: detection with gadoxetate disodium-enhanced MR imaging and multiphasic multidetector CT. Eur Radiol 22:845–854
Carbonell G, Kennedy P, Bane O et al (2022) Precision of MRI radiomics features in the liver and hepatocellular carcinoma. Eur Radiol 32:2030–2040
Fiset S, Welch ML, Weiss J et al (2019) Repeatability and reproducibility of MRI-based radiomic features in cervical cancer. Radiother Oncol 135:107–114
Zhao Y, Wu J, Zhang Q et al (2021) Radiomics analysis based on multiparametric MRI for predicting early recurrence in hepatocellular carcinoma after partial hepatectomy. J Magn Reson Imaging 53:1066–1079
Zhang Y, Lv X, Qiu J et al (2021) Deep learning with 3D convolutional neural network for noninvasive prediction of microvascular invasion in hepatocellular carcinoma. J Magn Reson Imaging 54:134–143
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Wei J, Jiang H, Zeng M et al (2021) Prediction of microvascular invasion in hepatocellular carcinoma via deep learning: a multi-center and prospective validation study. Cancers (Basel) 13:2368
Zhang XP, Chen ZH, Zhou TF et al (2019) A nomogram to predict early postoperative recurrence of hepatocellular carcinoma with portal vein tumour thrombus after R0 liver resection: a large-scale, multicenter study. Eur J Surg Oncol 45:1644–1651
Chan AWH, Zhong J, Berhane S et al (2018) Development of pre and post-operative models to predict early recurrence of hepatocellular carcinoma after surgical resection. J Hepatol 69:1284–1293
Zou ZM, Chang DH, Liu H, Xiao YD (2021) Current updates in machine learning in the prediction of therapeutic outcome of hepatocellular carcinoma: what should we know? Insights Imaging 12:31
Ahn JC, Connell A, Simonetto DA, Hughes C, Shah VH (2021) Application of artificial intelligence for the diagnosis and treatment of liver diseases. Hepatology 73:2546–2563
Zhang Z, Jiang H, Chen J et al (2019) Hepatocellular carcinoma: radiomics nomogram on gadoxetic acid-enhanced MR imaging for early postoperative recurrence prediction. Cancer Imaging 19:22
Kim S, Shin J, Kim DY, Choi GH, Kim MJ, Choi JY (2019) Radiomics on gadoxetic acid-enhanced magnetic resonance imaging for prediction of postoperative early and late recurrence of single hepatocellular carcinoma. Clin Cancer Res 25:3847–3855
Kooi T, Litjens G, van Ginneken B et al (2017) Large scale deep learning for computer aided detection of mammographic lesions. Med Image Anal 35:303–312
Hosny A, Parmar C, Coroller TP et al (2018) Deep learning for lung cancer prognostication: a retrospective multi-cohort radiomics study. PLoS Med 15:e1002711
Yang X, Kwitt R, Styner M, Niethammer M (2017) Quicksilver: fast predictive image registration - a deep learning approach. Neuroimage 158:378–396
Yasaka K, Akai H, Abe O, Kiryu S (2018) Deep learning with convolutional neural network for differentiation of liver masses at dynamic contrast-enhanced CT: a preliminary study. Radiology 286:887–896
Chaudhary K, Poirion OB, Lu L, Garmire LX (2018) Deep learning-based multi-omics integration robustly predicts survival in liver cancer. Clin Cancer Res 24:1248–1259
Kong C, Zhao Z, Chen W et al (2021) Prediction of tumor response via a pretreatment MRI radiomics-based nomogram in HCC treated with TACE. Eur Radiol 31:7500–7511
Song D, Wang Y, Wang W et al (2021) Using deep learning to predict microvascular invasion in hepatocellular carcinoma based on dynamic contrast-enhanced MRI combined with clinical parameters. J Cancer Res Clin Oncol 147:3757–3767
Zhang L, Xia W, Yan ZP et al (2020) Deep learning predicts overall survival of patients with unresectable hepatocellular carcinoma treated by transarterial chemoembolization plus sorafenib. Front Oncol 10:593292
Sung YS, Park B, Park HJ, Lee SS (2021) Radiomics and deep learning in liver diseases. J Gastroenterol Hepatol 36:561–568
Yang G, Nie P, Zhao L et al (2020) 2D and 3D texture analysis to predict lymphovascular invasion in lung adenocarcinoma. Eur J Radiol 129:109111
Shen C, Liu Z, Guan M et al (2017) 2D and 3D CT radiomics features prognostic performance comparison in non-small cell lung cancer. Transl Oncol 10:886–894
Friemel J, Rechsteiner M, Frick L et al (2015) Intratumor heterogeneity in hepatocellular carcinoma. Clin Cancer Res 21:1951–1961
Yang F, Wang Y, Li Q et al (2017) Intratumor heterogeneity predicts metastasis of triple-negative breast cancer. Carcinogenesis 38:900–909
Ji GW, Zhu FP, Xu Q et al (2019) Machine-learning analysis of contrast-enhanced CT radiomics predicts recurrence of hepatocellular carcinoma after resection: a multi-institutional study. EBioMedicine 50:156–165
Dai H, Lu M, Huang B et al (2021) Considerable effects of imaging sequences, feature extraction, feature selection, and classifiers on radiomics-based prediction of microvascular invasion in hepatocellular carcinoma using magnetic resonance imaging. Quant Imaging Med Surg 11:1836–1853
Ni M, Zhou X, Lv Q et al (2019) Radiomics models for diagnosing microvascular invasion in hepatocellular carcinoma: which model is the best model? Cancer Imaging 19:60
Acknowledgements
We sincerely thank Xiao Zhang in the First Affiliated Hospital of Jinan University and the Neusoft Research of Intelligent Healthcare Technology, Co. Ltd., for her assistance of deep learning analysis.
Funding
This study has received funding from the Science and Technology Planning Project of Guangdong Province of China (2017A020215010), Guangdong Basic and Applied Basic Research Foundation (2019A1515011269 and 2021A1515011305), and President Foundation of Zhujiang Hospital, Southern Medical University (yzjj2021qn23).
Author information
Authors and Affiliations
Corresponding authors
Ethics declarations
Guarantor
The scientific guarantor of this publication is Xianyue Quan.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethics approval
Institutional review board approval was obtained.
Methodology
• Retrospective
• Diagnostic or prognostic study
• Multicenter study
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Yan, M., Zhang, X., Zhang, B. et al. Deep learning nomogram based on Gd-EOB-DTPA MRI for predicting early recurrence in hepatocellular carcinoma after hepatectomy. Eur Radiol 33, 4949–4961 (2023). https://doi.org/10.1007/s00330-023-09419-0
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-023-09419-0