Abstract
Objective
To develop an automatic COVID-19 Reporting and Data System (CO-RADS)–based classification in a multi-demographic setting.
Methods
This multi-institutional review boards–approved retrospective study included 2720 chest CT scans (mean age, 58 years [range 18–100 years]) from Italian and Russian patients. Three board-certified radiologists from three countries assessed randomly selected subcohorts from each population and provided CO-RADS–based annotations. CT radiomic features were extracted from the selected subcohorts after preprocessing steps like lung lobe segmentation and automatic noise reduction. We compared three machine learning models, logistic regression (LR), multilayer perceptron (MLP), and random forest (RF) for the automated CO-RADS classification. Model evaluation was carried out in two scenarios, first, training on a mixed multi-demographic subcohort and testing on an independent hold-out dataset. In the second scenario, training was done on a single demography and externally validated on the other demography.
Results
The overall inter-observer agreement for the CO-RADS scoring between the radiologists was substantial (k = 0.80). Irrespective of the type of validation test scenario, suspected COVID-19 CT scans were identified with an accuracy of 84%. SHapley Additive exPlanations (SHAP) interpretation showed that the “wavelet_(LH)_GLCM_Imc1” feature had a positive impact on COVID prediction both with and without noise reduction. The application of noise reduction improved the overall performance between the classifiers for all types.
Conclusion
Using an automated model based on the COVID-19 Reporting and Data System (CO-RADS), we achieved clinically acceptable performance in a multi-demographic setting. This approach can serve as a standardized tool for automated COVID-19 assessment.
Keypoints
• Automatic CO-RADS scoring of large-scale multi-demographic chest CTs with mean AUC of 0.93 ± 0.04.
• Validation procedure resembles TRIPOD 2b and 3 categories, enhancing the quality of experimental design to test the cross-dataset domain shift between institutions aiding clinical integration.
• Identification of COVID-19 pneumonia in the presence of community-acquired pneumonia and other comorbidities with an AUC of 0.92.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Previously published articles have shown that COVID-19 has distinct imaging features making screening of suspected cases and evaluating disease progression using CT possible [1, 2]. Based on CT findings, several radiological societies have released standardized protocols for suspicion staging COVID-19 patients [3,4,5]. The COVID-19 Reporting and Data System or CO-RADS is a five-class suspicion classification scheme released by the Dutch Radiological Society (NVvR) [4]. CO-RADS includes discerning features related to unequivocal noninfectious origins, or community-acquired pneumonia from typical COVID-19 features on an unenhanced chest CT in a population with high incidence of COVID-19. This reporting system has shown high discriminatory power in triaging COVID-19 and provides an appropriate reporting language understandable by any radiologist [6, 7]. Therefore, using a standardized score such as CO-RADS is the most straightforward way of implementing an automated COVID-19 detection method [8].
The urgency to aid radiologists to detect COVID-19 has resulted in a rush to develop machine learning and deep learning models by neglecting a standardized approach. For example, the use of non-generalizable data for model training (single demographic, public datasets with different acquisition or reconstruction protocols, unavailability of source DICOM, scans with image artifacts) has resulted in poor application of machine learning methodology. This limitation has led to reproducibility issues and biases in recent study designs [9]. This can further aggravate as data characteristics change based on demographics, immunity landscape and clinical practice shift between different pandemic stages. Therefore, AI-driven studies should follow standardized and reproducible pathways to confirm the performance of AI models and their rapid adaptability and implementation into the clinical workflows.
Radiomics is a method of quantifying phenotypic characteristics of lesions in medical imaging using mathematical algorithms which can then be used to predict disease severity and progression [10, 11]. This quantitative process contrasts with the conventional radiological method, where the radiologist describes the lesions mainly based on qualitative attributes. The radiomic features are extracted at the sub-visual level, meaning that the computer system can detect patterns that might not be discernible by the human visual system. Therefore, when used in a standardized environment, radiomics may provide valuable clinical information complementary to conventional radiological analysis [12].
For the use of automated models in clinical practice, it is essential to consider three key aspects of model validation. The first aspect is to acquire high-quality data from a diverse population (multi-demographic) cohort. The second is to adopt a standardized annotation protocol understandable by radiologists. The third is to perform a thorough model analysis using various testing scenarios (internal–external validation). In our study, apart from including the aforementioned aspects, we performed model evaluation using datasets processed with and without noise reduction and interpreted the model output using radiomic features based on SHapley Additive exPlanations (SHAP) [13]. We hypothesize that in a multi-demographic setting, COVID-19 can be discerned automatically by predicting the CO-RADS score on chest CT using a classification algorithm combined with an optimal radiomic signature.
Materials and methods
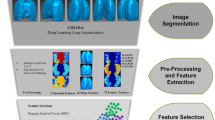
This retrospective study was approved by the ethics committees of the participating institutions. A graphical abstract of the workflow is shown in Fig. 1.
The overall workflow followed to validate machine learning models for automated COVID-19 suspicion staging based on the CO-RADS protocol. The scans with COVID-19 suspicion were selected from the Italian and Russian subcohorts retrospectively and annotated by experienced radiologists from different countries. In the first step, the datasets were processed using deep learning–based noise reduction (DLNR) and 3D segmentation masks were generated for each scan. Next, radiomic features were extracted and classified using ML algorithms. In the final step, statistical evaluation of the standard performance metrics and visualization of class-specific features were carried out to enhance the explainability of the models
Study population
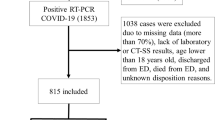
This study included 1418 chest CTs from patients suspected of having COVID-19 from two countries: the first population, from MosMed in Moscow, Russia, collected between March and April 2020 and the second population from the COVID center of Università Campus Bio-Medico di Roma, Rome, Italy, collected between February and May 2020 [14, 15]. Although the acquisition protocol was entirely different for both populations, the received data set primarily consisted of anonymized unenhanced chest CT imaging with an average of 300–400 images (slices) per patient scan. The inclusion and exclusion criteria of the acquired dataset are shown in Fig. 2, and the overview of the acquisition and reconstruction parameters is shown in Table 1.
Data flowchart of Russian and Italian subcohorts included for the study with the training and test split. Below the flowchart, a description of different evaluation settings of data scenarios is depicted. Note that n refers to the number of patients. CO-RADS–COVID-19 Reporting and Data Systems, CT 0, 1, 2, 3, 4–severity–based Russian annotations
Radiologists’ annotation
To explicate the acquired datasets, three board-certified multi-demographic radiologists (Electronic supplementary material, section 1) with more than 5 years of experience reviewed the CT images from multi-demographic datasets. The CO-RADS annotation protocol was followed by each radiologist to capture the multi-class variation in COVID-19 suspected candidates. The readers were blinded to the clinical–epidemiological diagnosis of COVID-19 and the predefined CT category (severity levels) that existed in the Russian dataset. In the given time, we were able to retrieve systematic annotations for 673 patient scans. An overlap subset of 43 patients was selected randomly from the included population to calculate the inter-observer agreement between multi-demographic radiologists. The CT scans were viewed using an open-source Multi-image Analysis GUI [16], with readers being able to modify the window settings.
Noise reduction
In our study, we used a proprietary deep learning–based noise reduction (DLNR) algorithm (Pixelshine, AlgoMedica) which has been shown to effectively reduce image noise of an already reconstructed DICOM image and can improve the image quality [17,18,19]. All the scans irrespective of CT protocol were processed using the DLNR algorithm for analysis. In total, two subsets of data were created, one without noise reduction (setting 1, 3, and 5) and one with noise reduction (setting 2, 4, and 6). The reason for using noise reduction is to ensure a more consistent presentation of the data obtained with different protocols.
Feature extraction
After 3D lung lobe segmentation (Electronic supplementary material, section 2) of each scan, radiomic features were derived using the PyRadiomics library [20]. Most of the feature descriptors extracted using PyRadiomics are in compliance with feature definitions described by the Imaging Biomarker Standardization Initiative (IBSI) [21]. Additionally, 26 new radiomic descriptors known as co-occurrence of local anisotropic gradient orientations (CoLlAGe) features were included to capture the lobe-level anisotropy and encapsulate CO-RADS [22].
Furthermore, to increase the feature diversity, features were extracted after applying filters such as Wavelet, Laplacian of Gaussian (LoG), Square, Square Root, Logarithmic, Exponential, Gradient, and Local Binary Pattern 2D (LBP2D) to the original image. However, filters were not applied to the two-dimensional shape–based category and CoLlAGe features.
Experimental design and performance evaluation
We used the machine learning (ML) models logistic regression (LR), multilayer perceptron (MLP), and random forest (RF) for automated CO-RADS classification. For comparison purposes, we evaluated the ML models under different combinations of CO-RADS operating points as described recently [8]. To account for the generalizability and reproducibility using multi-demographic datasets in the three machine learning models, we considered two scenarios for model evaluation. In the first scenario, we trained the classification models on a mixed dataset (Russian and Italian datasets) and tested on an internal hold-out dataset (Russian). In the second scenario, the classification models were trained either on the Russian or on the Italian dataset and externally validated on the other population. Since these scenarios were evaluated with and without noise reduction for classification, in total, we implemented six different settings to thoroughly evaluate all the possibilities of model training. The details of each setting including the data split are shown in Fig. 2.
Statistics and evaluation metrics
To understand the inter-observer variability in CO-RADS annotation between the radiologists, we used Fleiss kappa and t test.
Before model training, the extracted features were scaled to the same bandwidth using Z-score normalization. We used the Pearson’s correlation score (PCC), to eliminate the redundant features with PCC greater than 0.95. As a feature selection method, we used one-way ANOVA F-test and features with a significant difference (p < 0.05) between the groups were considered [23]. This feature scaling and feature selection application is referred to as feature engineering in the rest of the paper. To avoid over-fitting due to class imbalance that existed in CO-RADS classes, we used the Synthetic Minority Oversampling Technique (SMOTE) [24].
In our experiments, we calculated the area under the receiver operation curve (AUC) to understand the performance of ML–based automatic CO-RADS scoring along with F1 measure, recall, accuracy, and precision. DeLong’s test was used to compare the statistical difference between AUC [25]. The 95% confidence interval (95% CI) for the metrics was evaluated using non-parametric bootstrapping with 1000 iterations. The complete experimental setup was built in-house using the PyTorch framework in Python [26].
Explainable visualizations
For better understanding of model prediction, we used SHapley Additive exPlanations (SHAP) which measures the feature importance based on the model output [27]. The SHAP five-class plot was used to visualize feature importance on the CO-RADS classification for the best model. The independent class summary plot for COVID-19 (CO-RADS 5) was used to understand the relationship between the feature and the impact on the prediction.
Results
Patient characteristics
We excluded 15 scans for which CO-RADS grade 0 (not interpretable) was assigned. In total, 34.5% (227/658) of the patients had no COVID-19 CT findings (CO-RADS 1) or normal, and 6.8% (45/658) of the patients had CT findings consistent with infections other than COVID-19 (CO-RADS 2), which mainly included multiple nodules, emphysema, fibrosis, bronchiectasis, calcification, lung abscess, tree-in-bud patterns, and pulmonary metastasis. About 17.2% (113/658) of the patients who had CT findings compatible with COVID-19, but also other disease like bacterial infection (aspecific GGOs, consolidations, pleural fluids), were categorized as CO-RADS 3. The CO-RADS 4 category contained 16.6% (109/658) of the patients who were suspected with COVID-19 abnormalities, and 24.9% (164/658) of the patients who had confirmed COVID-19 infection were categorized as CO-RADS 5 (Table 2). In the confirmed cases of both the subcohorts, more than 50% (194/297, 200/361) of the cases were found to have ground glass opacities. Other commonly observed findings were pleural effusion, consolidation, crazy paving, and vascular thickening.
Inter-observer agreement of multi-demographic radiologist CO-RADS grading systems
For overall CO-RADS annotation, we observed substantial agreement between the radiologists with a mean Fleiss’ kappa (k) of 0.802 (95% CI: 0.705–0.899). For individual CO-RADS class, between the readers, there was almost perfect agreement for CO-RADS 5 (k = 1.00, 95% CI: 0.84–1.17), CO-RADS 1 (k = 0.93, 95% CI: 0.76–1.10), and CO-RADS 3 (k = 0.91, 95% CI: 0.74–1.00). Substantial agreement was observed for CO-RADS 2 and CO-RADS 4 with a k value of 0.76 (95% CI: 0.58–0.93) and 0.72 (95% CI: 0.56–0.92) respectively (Table 3).
Performance of models in different data settings
In this section, we report mean AUC of each classifier for overall CO-RADS classification and the percentage change in performance after noise reduction.
The logistic regression (LR) classifier, in setting 1, performed with a mean AUC of 0.83 ± 0.01 for overall CO-RADS classification and 0.83 ± 0.025 after noise reduction (setting 2). In settings 4 and 6 (after noise reduction), the LR classifier performance increased by 4% when compared to settings 3 and 5; that is, AUC increased from 0.71 ± 0.06 to 0.74 ± 0.04 and from 0.70 ± 0.02 to 0.75 ± 0.03 respectively.
The multilayer perceptron (MLP) classifier in setting 1 and setting 2 performed with an AUC of 0.84 ± 0.02 and 0.87 ± 0.06 respectively. In setting 3, the AUC was 0.72 ± 0.02, and in setting 4, the AUC was 0.77 ± 0.04. In setting 4, the AUC was 0.69 ± 0.021, and in setting 5, the AUC was 0.75 ± 0.01. Hence, after noise reduction, we observed 3%, 5%, and 6% increase in the performance of the MLP classifier in settings 2, 4, and 6 respectively.
The random forest model had an AUC of 0.88 ± 0.02 in setting 1 and 0.93 ± 0.04 in setting 2. In setting 3, the AUC was 0.76 ± 0.04, and in setting 4, the AUC was 0.78 ± 0.04. In setting 5, the AUC was 0.70 ± 0.02, and in setting 6, the AUC was 0.77 ± 0.02. Hence, after noise reduction, we observed 5%, 2%, and 7% increase in performance settings 2, 4, and 6 respectively. RF classifier outperformed other classifiers in most of the settings.
The details of the CO-RADS score of all combinations for COVID predictions are shown in Table 4, and the other evaluation metrics are shown in Table 5. It can be observed that the performance of all the three classifiers in settings 1 and 2 was better than that of the rest of the settings. Additionally, we investigated the model capability for prediction of “normal chest CT” both with and without noise reduction and the plot is shown in Fig. 3. Among all the three ML classifiers, random forest performed the best in almost all settings.
COVID-19 prediction
Here, we present the results for COVID-19 prediction (CO-RADS 5) for the best classifier in each setting. In setting 1, on the hold-out dataset, the random forest (RF) classifier classified patients with COVID-19 with an AUC of 0.88 ± 0.06. The sensitivity and specificity in identifying COVID-19 features for the RF algorithm was 0.86 ± 0.11 and 0.88 ± 0.06 respectively. After noise reduction, the performance of the RF model increased to AUC of 0.92 ± 0.06 with sensitivity = 0.91 ± 0.07 and specificity = 0.94 ± 0.04.
We observed a performance drop in the RF model for scenario 2, when trained on the Russian dataset and externally validated on the Italian dataset, and vice versa. The RF classifier achieved an AUC of 0.79 ± 0.07 without any noise reduction. The sensitivity and specificity of the RF model was 0.72 ± 0.10 and 0.73 ± 0.05 respectively. In setting 4, with noise reduction, AUC was 0.75 ± 0.08, sensitivity = 0.95 ± 0.03, and specificity = 0.63 ± 0.03 for COVID-19 prediction.
For setting 5 of scenario 2, the optimal model performance was observed with an AUC of 0.71 ± 0.06, sensitivity = 0.74 ± 0.09, and specificity = 0.66 ± 0.05. After noise reduction, the optimal model performance increased to 0.75 ± 0.09 (AUC), sensitivity = 0.75 ± 0.09, and specificity = 0.75 ± 0.05. The detailed evaluation metrics of the RF classifier in each setting are shown in Table 4.
Interpretation of radiomic signature
To recognize the important features in each scenario for CO-RADS classification, we visualized the SHAP values of the top 20 features. The SHAP values of each setting are summarized in Fig. 4.
Top twenty feature visualization using SHAP for CO-RADS classification in each data setting. The features are arranged in a descending order of feature importance (SHAP values). Using this feature importance map, one can observe how each feature contributes to the machine learning model’s predictions and identifies the common features
Out of the top 20 features responsible for CO-RADS classification of scenario 1, we found “log (sigma=1)-3D-GLCM Auto-correlation,” “log (sigma=1) 3D GLCM Joint Entropy,” “log (sigma=1) 3D GLCM Idm,” “log(sigma=1) 3D GLDM Low Gray Level Emphasis,” and “original Firstorder-10Percentile” features to be the most common in settings 1 and 2. Similarly, for other settings, important CO-RADS classification features can be realized in Fig. 4.
The individual summary plot showed class-specific features for CO-RADS 5 (Fig. 5). For each setting, a positive SHAP value indicates an increase in the risk of COVID-19. “Log (sigma=1) GLDM Small Dependence Low Gray Level Emphasis” showed negative impact, and “wavelet (LH) GLCM Imc1” showed positive impact on COVID prediction in almost all settings.
Discussion
Several deep learning [28, 29], radiomic [30, 31], and integrated models [32, 33] have been developed since the outbreak of COVID-19, focusing on screening, diagnosis, and prognosis of COVID-19. To facilitate the translation of COVID-19 detection models into clinical practice, we followed a systematic approach to develop simple, generalizable, and reproducible automated COVID-19 detection models. In this study, (A) we successfully evaluated multi-site or public–private dataset scenarios and showed overall CO-RADS classification improvement by using noise reduction technique; (B) by choosing high-quality data from two clinical centers and heterogenous imaging protocols, we significantly reduced the population selection bias encountered in many recent studies as mentioned in a recent review [9]; (C) the annotation was done by three experienced radiologists from different countries avoiding the annotation bias that can occur when using inexperienced radiologists or radiologists from the same site; (D) by using a standardized annotation protocol, we made our study easy to implement and compare; (E) the interpretation of radiomic signatures via SHAP enabled us to pinpoint the key features that influence classification; and (F) finally, by carrying out validation procedures resembling the guidelines listed in category 2b and 3 of the Transparent Reporting of a multivariable prediction model for Individual Prognosis or Diagnosis (TRIPOD) statement, we have enhanced the quality of experimental design, and demonstrated the usefulness of the prediction models by testing the cross-dataset domain shift between different institutions [34].
Regardless of the type of validation scenario, CT radiomic features facilitated the identification of CT scans with suspected COVID-19 with accuracy of 84% (138/164) in a two-dimensional setting. Homayounieh et al, Fang et al, and Liu et al reported higher accuracies using a combined clinical–radiological radiomic signature, with their training based solely on a single demographic group, which may raise concerns of bias [30, 31, 35]. Lee et al used a deep learning algorithm and achieved accuracies similar to ours while investigating a heterogeneous population from more than eight countries [36].
We observed that among the two validation methods, the internal hold-out method had the highest AUC for COVID classification (CO-RADS 5) with 0.92 compared to the external validation (0.75). This shows that the model trained on multi-demographics and tested on internal hold-out performs better when compared to the model trained on single demographic data [36]. There have been many automatic models to differentiate COVID-19 from non-COVID-19 but a very few that have the capability to segregate comorbidities such as emphysema and lung cancer [2]. Notably, our multi-class classification model (RF) was able to identify other comorbidities with the highest accuracy of 85% (329/386). Prior works have shown that radiomics can be used to distinguish COVID-19 (CO-RADS 4) from other pneumonias (CO-RADS 3) on CT [37, 38]. We observed an average performance of 0.80 (AUC) in differentiating other community-acquired pneumonias (CO-RADS 3) in patients with suspected COVID-19, and in contrast to previous works, we used scans from patients with suspected COVID-19 without including scans from the general population.
Studies have shown the importance of using whole lung CT radiomics for predicting outcome and disease severity of COVID-19 patients compared with subjective radiologist assessment [30, 31, 39, 40]. Although the direct comparison of the features obtained in these studies cannot be compared to our study, we observed that textural features dominated feature importance for COVID-19 patients or CO-RADS 5 (Fig. 5). Interestingly, by employing noise reduction on non-contrast chest CTs, we were able to improve the performance for all the classifiers. This could be because apart from reducing noise and aiding dimensionality reduction of radiomic features, it helped in generalizing the scans acquired from different acquisition protocols (minimizing the variance).
Although the radiologists were from different countries (The Netherlands, Germany, and Italy), inter-observer agreement in CO-RADS scoring in multi-demographic data was substantial. Similar agreement was reported by Prokop et al, Lessmann et al, and Dilek et al [4, 7, 8].
Some of the evident limitations of this study are that our radiomic signature could also include epidemiological–laboratory parameters and clinical symptoms to provide a more comprehensive model. Secondly, it is a well-known fact that variation in the acquisition process, reconstruction parameters, or study protocol can result in different radiomic features. The degree to which this variation affects classification performance is an area of active research. The third limitation is that only non-contrast images were included. However, comparing and combining features engineered by deep learning approach would be the primary focus of our future work [41, 42].
Conclusions
We have attempted to answer the question of whether we can use a systematic approach to construct a radiomic signature and employ simple ML classifiers that can effectively distinguish COVID-19 in a multi-demographic setting. The best classifier on average correctly designates the CO-RADS score in 80% of the cases. That is, by harnessing the power of radiomics combined with noise reduction, it is possible to predict the CO-RADS score from a non-contrast chest CT with a relatively high accuracy. Adopting the aforementioned model into clinical practice as a standardized tool may aid radiologists in classifying COVID-19. Lastly, this study design can be used as a research tool, facilitating reproducible and comparable models in the field of automated COVID-19 detection.
Abbreviations
- AUC:
-
Area under the receiver operation curve
- CoLliAGe:
-
Co-occurrence of local anisotropic gradient orientations
- CO-RADS:
-
COVID-19 Reporting and Data System
- CT:
-
Computed tomography
- COVID-19:
-
Coronavirus disease 2019
- DLNR:
-
Deep learning–based noise reduction
- GLCM:
-
Gray-level co-occurrence matrix
- GLDM:
-
Gray-level dependence matrix
- GLRLM:
-
Gray-level run length matrix
- GLSZM:
-
Gray-level size zone matrix
- LBP2D:
-
Local binary pattern 2D
- LoG:
-
Laplacian of Gaussian
- LR:
-
Logistic regression
- MLP:
-
Multilayer perceptron
- NR:
-
Noise reduction
- RF:
-
Random forest
- RT-PCR:
-
Reverse transcription–polymerase chain reaction
- SHAP:
-
SHapley Additive exPlanations
References
Byrne D, Neill SBO, Müller NL et al (2021) RSNA expert consensus statement on reporting chest CT findings related to COVID-19: interobserver agreement between chest radiologists. Can Assoc Radiol J 72:159–166
Dong D, Tang Z, Wang S et al (2021) The role of imaging in the detection and management of COVID-19: a review. IEEE Rev Biomed Eng 14:16–29. https://doi.org/10.1109/RBME.2020.2990959
He Y (2020) Translation: diagnosis and treatment protocol for novel coronavirus pneumonia (Trial Version 7). Infect Microbes Dis 2:48–54. https://doi.org/10.1097/IM9.0000000000000022
Prokop M, van Everdingen W, van Rees VT et al (2020) CO-RADS: a categorical CT assessment scheme for patients suspected of having COVID-19—definition and evaluation. Radiology 296:E97–E104. https://doi.org/10.1148/radiol.2020201473
Simpson S, Kay FU, Abbara S et al (2020) Radiological Society of North America expert consensus document on reporting chest CT findings related to COVID-19: endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. Radiol Cardiothorac Imaging 2:e200152. https://doi.org/10.1148/ryct.2020200152
de Smet K, de Smet D, Demedts I et al (2020) Diagnostic power of chest CT for COVID-19: to screen or not to screen. medArxiv. https://doi.org/10.1101/2020.05.18.20097444
Dilek O, Kaya O, Akkaya H et al (2021) Diagnostic performance and interobserver agreement of CO-RADS: evaluation of classification in radiology practice. Diagn Interv Radiol 27:615–620. https://doi.org/10.5152/dir.2021.201032
Lessmann N, Sánchez CI, Beenen L et al (2021) Automated assessment of COVID-19 reporting and data system and chest CT severity scores in patients suspected of having COVID-19 using artificial intelligence. Radiology 298:E18–E28. https://doi.org/10.1148/radiol.2020202439
Roberts M, Driggs D, Thorpe M et al (2021) Common pitfalls and recommendations for using machine learning to detect and prognosticate for COVID-19 using chest radiographs and CT scans. Nat Mach Intell 3:199–217
Larue RTHM, Defraene G, De Ruysscher D et al (2017) Quantitative radiomics studies for tissue characterization: a review of technology and methodological procedures. Br J Radiol 90:20160665. https://doi.org/10.1259/bjr.20160665
van Ooijen PMA, Nagaraj Y, Olthof A (2020) Medical imaging informatics, more than ‘just’ deep learning. Eur Radiol 30:5507–5509. https://doi.org/10.1007/s00330-020-06939-x
Lambin P, Leijenaar RTH, Deist TM et al (2017) Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol 14:749–762
Lundberg S, Lee S-I (2017) A unified approach to interpreting model predictions. In: Proceedings of the 31st international conference on neural information processing systems. Curran Associates Inc., Red Hook, NY, USA, pp 4768–4777
Morozov SP, Andreychenko AE, Blokhin IA et al (2020) MosMedData: data set of 1110 chest CT scans performed during the COVID-19 epidemic. Digit Diagnostics 1:49–59. https://doi.org/10.17816/DD46826
Quattrocchi CC, Mallio CA, Presti G et al (2020) The challenge of COVID-19 low disease prevalence for artificial intelligence models: report of 1,610 patients. Quant Imaging Med Surg 10:1891–1893. https://doi.org/10.21037/qims-20-782
Lancaster L, Martinez MJ (2021) Multi-image analysis GUI Viewer.4.0.1. http://ric.uthscsa.edu/mango/index.html. Accessed November 18, 2016
Tian S, Liu A, Liu J et al (2019) Potential value of the PixelShine deep learning algorithm for increasing quality of 70 kVp+ASiR-V reconstruction pelvic arterial phase CT images. Jpn J Radiol 37:186–190. https://doi.org/10.1007/s11604-018-0798-0
Wisselink HJ, Pelgrim GJ, Rook M et al (2020) Potential for dose reduction in CT emphysema densitometry with post-scan noise reduction: a phantom study. Br J Radiol 93:20181019. https://doi.org/10.1259/bjr.20181019
Steuwe A, Weber M, Bethge OT et al (2021) Influence of a novel deep-learning based reconstruction software on the objective and subjective image quality in low-dose abdominal computed tomography. Br J Radiol 94:20200677
Van Griethuysen JJM, Fedorov A, Parmar C et al (2017) Computational radiomics system to decode the radiographic phenotype. Cancer Res 77:e104–e107
Zwanenburg A, Leger S, Vallières M et al (2016) Image biomarker standardisation initiative-feature definitions. arXiv Prepr arXiv161207003
Prasanna P, Tiwari P, Madabhushi A (2016) Co-occurrence of local anisotropic gradient orientations (CoLlAGe): a new radiomics descriptor. Sci Rep 6:37241
Higuchi K, Nagao M, Matsuo Y et al (2013) Detection of ground-glass opacities by use of hybrid iterative reconstruction (iDose) and low-dose 256-section computed tomography: a phantom study. Radiol Phys Technol. https://doi.org/10.1007/s12194-013-0200-y
Chawla NV, Bowyer KW, Hall LO, Kegelmeyer WP (2002) SMOTE: synthetic minority over-sampling technique. J Artif Intell Res 16:321–357
DeLong ER, DeLong DM, Clarke-Pearson DL (1988) Comparing the areas under two or more correlated receiver operating characteristic curves: a nonparametric approach. Biometrics 44(3):837–845
Paszke A, Gross S, Massa F et al (2019) PyTorch: an imperative style, high-performance deep learning library. In: Advances in neural information processing systems 32. Curran Associates, Inc., pp 8024–8035
Lundberg SM, Nair B, Vavilala MS et al (2018) Explainable machine-learning predictions for the prevention of hypoxaemia during surgery. Nat Biomed Eng 2:749–760
Shoeibi A, Khodatars M, Alizadehsani R, et al (2020) Automated detection and forecasting of covid-19 using deep learning techniques: a review. arXiv Prepr arXiv200710785
Chen Y, Jiang G, Li Y et al (2020) A survey on artificial intelligence in chest imaging of COVID-19. Bio Integr 1:137–146
Fang X, Li X, Bian Y et al (2020) Radiomics nomogram for the prediction of 2019 novel coronavirus pneumonia caused by SARS-CoV-2. Eur Radiol 30:6888–6901. https://doi.org/10.1007/s00330-020-07032-z
Homayounieh F, Ebrahimian S, Babaei R et al (2020) CT radiomics, radiologists, and clinical information in predicting outcome of patients with COVID-19 pneumonia. Radiol Cardiothorac Imaging 2:e200322. https://doi.org/10.1148/ryct.2020200322
Wang H, Wang L, Lee EH et al (2021) Decoding COVID-19 pneumonia: comparison of deep learning and radiomics CT image signatures. Eur J Nucl Med Mol Imaging 48:1478–1486. https://doi.org/10.1007/s00259-020-05075-4
Zhang X, Wang D, Shao J et al (2021) A deep learning integrated radiomics model for identification of coronavirus disease 2019 using computed tomography. Sci Rep 11:3938. https://doi.org/10.1038/s41598-021-83237-6
Collins GS, Reitsma JB, Altman DG, Moons K (2015) Transparent reporting of a multivariable prediction model for individual prognosis or diagnosis (TRIPOD): the TRIPOD statement. BMC Med 13:1. https://doi.org/10.1186/s12916-014-0241-z
Liu H, Ren H, Wu Z et al (2021) CT radiomics facilitates more accurate diagnosis of COVID-19 pneumonia: compared with CO-RADS. J Transl Med 19:29. https://doi.org/10.1186/s12967-020-02692-3
Lee EH, Zheng J, Colak E et al (2021) Deep COVID DeteCT: an international experience on COVID-19 lung detection and prognosis using chest CT. NPJ Digit Med 4:11. https://doi.org/10.1038/s41746-020-00369-1
Li L, Qin L, Xu Z et al (2020) Using artificial intelligence to detect COVID-19 and community-acquired pneumonia based on pulmonary CT: evaluation of the diagnostic accuracy. Radiology 296:E65–E71. https://doi.org/10.1148/radiol.2020200905
Pu J, Leader J, Bandos A et al (2020) Any unique image biomarkers associated with COVID-19? Eur Radiol 30:6221–6227. https://doi.org/10.1007/s00330-020-06956-w
Guiot J, Vaidyanathan A, Deprez L et al (2020) Development and validation of an automated radiomic CT signature for detecting COVID-19. Diagnostics 11:41. https://doi.org/10.3390/diagnostics11010041
Xie Z, Sun H, Wang J et al (2021) A novel CT-based radiomics in the distinction of severity of coronavirus disease 2019 (COVID-19) pneumonia. BMC Infect Dis 21:608. https://doi.org/10.1186/s12879-021-06331-0
Chao H, Fang X, Zhang J et al (2021) Integrative analysis for COVID-19 patient outcome prediction. Med Image Anal 67:101844
Avanzo M, Gagliardi V, Stancanello J et al (2021) Combining computed tomography and biologically effective dose in radiomics and deep learning improves prediction of tumor response to robotic lung stereotactic body radiation therapy. Med Phys 48:6257–6269. https://doi.org/10.1002/mp.15178
Acknowledgements
We gratefully acknowledge the support of NVIDIA Corporation with the donation of the Titan XP GPU used for this research.
Funding
Part of this work was realized within the DAME-project, funded by the INTERREG V A- Deutschland-Nederland program with resources from the European Regional Development Fund and co-funded by the Ministerie van Economische Zaken en Klimaat (EZK), the Province of Groningen, and the Niedersächsisches Ministerium für Bundes- und Europaangelegenheiten und Regionale Entwicklung. A. Andreychenko, N. Pavlov, and S. Morozov received funding as part of the research (No. in the Unified State Information System for Accounting of Research, Development, and Technological Works (EGISU): АААА-А20-120071090056-3) under the Program of the Moscow Healthcare Department “Scientific Support of the Capital's Healthcare” for 2020–2022.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is dr. ir. PMA (Peter) van Ooijen.
Conflict of Interest
Matthijs Oudkerk holds a financial interest in the Institute of DiagNostic Accuracy Research (iDNA), an organization that aims to speed up the global implementation of the early detection of lung cancer with comorbidities in cardiovascular diseases and COPD.
The rest of the authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and Biometry
Three of the authors have significant statistical expertise.
Informed Consent
Written informed consent was obtained from all subjects of the Italian and Russian study.
Ethical Approval
This retrospective study was approved by the ethics committees of the participating institutions.
Methodology
• Retrospective study
• Diagnostic study
• Multicenter study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 18 kb)
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Nagaraj, Y., de Jonge, G., Andreychenko, A. et al. Facilitating standardized COVID-19 suspicion prediction based on computed tomography radiomics in a multi-demographic setting. Eur Radiol 32, 6384–6396 (2022). https://doi.org/10.1007/s00330-022-08730-6
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-022-08730-6