Abstract
Objectives
Based on the current clinical routine, we aimed to develop a novel deep learning model to distinguish coronavirus disease 2019 (COVID-19) pneumonia from other types of pneumonia and validate it with a real-world dataset (RWD).
Methods
A total of 563 chest CT scans of 380 patients (227/380 were diagnosed with COVID-19 pneumonia) from 5 hospitals were collected to train our deep learning (DL) model. Lung regions were extracted by U-net, then transformed and fed to pre-trained ResNet-50-based IDANNet (Identification and Analysis of New covid-19 Net) to produce a diagnostic probability. Fivefold cross-validation was employed to validate the application of our model. Another 318 scans of 316 patients (243/316 were diagnosed with COVID-19 pneumonia) from 2 other hospitals were enrolled prospectively as the RWDs to testify our DL model’s performance and compared it with that from 3 experienced radiologists.
Results
A three-dimensional DL model was successfully established. The diagnostic threshold to differentiate COVID-19 and non-COVID-19 pneumonia was 0.685 with an AUC of 0.906 (95% CI: 0.886–0.913) in the internal validation group. In the RWD cohort, our model achieved an AUC of 0.868 (95% CI: 0.851–0.876) with the sensitivity of 0.811 and the specificity of 0.822, non-inferior to the performance of 3 experienced radiologists, suggesting promising clinical practical usage.
Conclusions
The established DL model was able to achieve accurate identification of COVID-19 pneumonia from other suspected ones in the real-world situation, which could become a reliable tool in clinical routine.
Key Points
• In an internal validation set, our DL model achieved the best performance to differentiate COVID-19 from non-COVID-19 pneumonia with a sensitivity of 0.836, a specificity of 0.800, and an AUC of 0.906 (95% CI: 0.886–0.913) when the threshold was set at 0.685.
• In the prospective RWD cohort, our DL diagnostic model achieved a sensitivity of 0.811, a specificity of 0.822, and AUC of 0.868 (95% CI: 0.851–0.876), non-inferior to the performance of 3 experienced radiologists.
• The attention heatmaps were fully generated by the model without additional manual annotation and the attention regions were highly aligned with the ROIs acquired by human radiologists for diagnosis.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
The newly emerging coronavirus disease (COVID-19, named by WHO) has spread globally and brought about 165,000 deaths and huge economic loss [1, 2]. This is the third zoonotic coronavirus breakout in the twenty-first century and has become a daunting challenge to human beings [3]. With the rapid spread in a variety of countries, new requirements for epidemic prevention and control are put forward [4,5,6]. Nowadays, the diagnosis of COVID-19 totally depends on a SARS-CoV-2 virus–specific reverse transcriptase-polymerase chain reaction (RT-PCR) test. New methods were developed or under development [7,8,9]. Chest computed tomography (CT) is important in the diagnosis and treatment of lung diseases including viral pneumonia. Compared with molecular diagnostic testing, CT scanning has the advantages of a faster turnaround time, more detailed information related to pathology, and quantitative measurement of lesion size and lung involvement, which may have important implications for prognosis [10].
The subpleural distributed ground-glass opacities (GGOs) and “crazy paving” signs were reported by several papers to be the typical findings in COVID-19 pneumonia patients [10, 11]. However, there are no unique manifestations of COVID-19 pneumonia on CT scans. Although the Fleischner Society has published a guideline to help radiologists identify the typical features of COVID-19 pneumonia, so far there are no high-level evidence-based diagnostic tests to clarify the diagnostic efficiency of such features acquired by radiologists [12]. These non-quantifiable radiological findings were too subjective to establish a diagnostic criterion of COVID-19 pneumonia based on human-perceived CT findings [13, 14].
In recent years, deep learning (DL) has exhibited promising potential in automatic diagnosis and differential diagnosis of various diseases [15,16,17]. There have been lots of studies which take advantage of convolutional neural network (CNN) to solve medical problems, such as pneumonia detection and classification, and have outperformed not only the traditional machine learning but also human benchmarks applied in previous studies [15,16,17,18,19]. Several new DL models have been developed to make an accurate diagnosis of COVID-19 pneumonia based on chest CT images [20,21,22]. However, few prospective deep learning studies or randomized trials exist in this field. Most independent datasets to test DL models are likely to have a high risk of bias [23]. It is important to validate the generalization ability of DL models by real-world dataset (RWD) which could really help to realize the transformation from the academy to clinical practice [24, 25].
In this study, we attempted to construct a novel deep learning model to distinguish COVID-19 pneumonia from all suspected COVID-19 pneumonia and validated it with an RWD to testify its application value in clinical routine.
Materials and methods
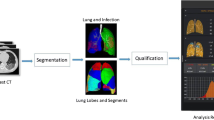
Our institutional review board approved this multi-center retrospective study and waived the requirement of written informed consent. De-identified data were used to prevent any leak of patient’s privacy. The workflow is depicted in Fig. 1.
Patient characteristics for model-training group
To establish our artificial intelligence COVID-19 classification model, from Jan. 1 to March 18, 2020, 563 chest CT exams from 380 patients were enrolled in the model-training group. CT scans were selected from 5 institutions in Anhui, Zhejiang Province, and Shanghai which met the following criteria: (1) suspected viral pneumonia manifestations presented on chest CT scans including single or scattered GGO or GGO-predominant density, (2) laboratory tests and RT-PCR tests were taken to clarify the pathogen of pneumonia, (3) no significant artifacts observed. Fivefold cross-validation was used for hyperparameter fine-tuning and model evaluation.
Patient characteristics for real-world data
To address regional variations and general applicability of our DL diagnostic model, the performance was tested in a real-world cohort from two institutions in a prospective fashion: one from the epicenter Hubei, China (City of Wuhan), and the other from the non-epidemic areas in China (City of Shanghai).
The inclusion criteria for the RWD cohort were listed as follows: (1) suspected COVID-19 manifestations presented on chest CT scans including GGO or GGO-predominant density; (2) no significant artifacts observed. After being reported as suspected COVID-19 by radiologists, these patients were visited by epidemiologists in the hospital based on clinical information, laboratory, and radiological results, then RT-PCR tests were taken for final diagnosis. We consecutively enrolled patients (n = 3416) who took CT scans in INSTITUTION ONE (Huashan Hospital, representing non-epidemic area) from Jan 11 to April 11, 2020, and all patients (n = 328) who took CT scan in INSTITUTION TWO (Wuhan Fangcang Hospital, representing epidemic area) from Feb. 21 to March 8, 2020. Among them, a total of 316 patients met our criteria and were consecutively enrolled in our RWD.
CT scanning protocol
A total of 54 CT scans of 52 patients from Institution train-A (Huashan North Hospital) were imaged with a 16-section CT scanner (uCT 510, UIH). Six CT scans of 6 patients from Institution train-B (Taizhou People’s Hospital) were imaged with a 16-section CT scanner (LightSpeed CT, GE Medical System). A total of 58 CT scans of 58 patients from Institution train-C (Huashan East Hospital) were imaged with a 64-section CT scanner (Aquilion Prime, Toshiba Medical Systems). In total, 375 CT scans of 197 patients from Institution train-D (Fuyang No. 2 People’s Hospital) were imaged with a 64-section CT scanner (Aquilion 64, Toshiba Medical Systems). Seventy CT scans of 70 patients from Institution train-E (Ma’Anshan No. 4 People’s Hospital) were imaged with a 64-section CT scanner (Siemens Somatom Sensation). A total of 85 scans of 83 patients from Institution test-A were imaged with a 64-section CT scanner (Discovery CT, GE Medical System). A total of 233 scans of 233 cases from Institution test-B were imaged with a 16-section CT scanner (uCT 550, UIH, China). Images were photographed at the lung (window width, 1500 HU; window level, − 500 HU) and mediastinal (window width, 320 HU; window level, 40 HU) windows with 5-mm thickness.
Deep learning model
We utilized a 3D DL framework to distinguish COVID-19 from other suspected viral pneumonia by clinicians, referring to IDANNet. It could effectively extract 2D local features and 3D global features. The IDANNet used ResNet50 as the backbone to take CT slices as input and extract features for each slice. Then, the extracted slice features were fed into a feature fusion layer to capture sequence dependency following a max-pooling layer. The feature fusion layer consisted of two-layer CNN. The final extracted features were fed into a dense layer following SoftMax activation to generate the probability for COVID-19 pneumonia (Fig. 2).
The illustration of the network architectures of our proposed deep learning (DL) model, including U-net and COVIDNet. a U-net is composed of a two-stage segmentation module for acceleration. In the first stage, we down-sampled the input image to a 128 × 128 level and segmented the lung field from the image, as the patterns of lung fields were easily learned at a relatively low resolution. In the second stage, we first calculated the bounding box with the lung field segmentation results. The key region was cropped from the original input image and resized it to a 256 × 256 level as the input for the second stage segmentation model. b The 3D classification networks (COVIDNet) were used in our COVID-19 diagnosis system. It is a convolutional neural network using ResNet50 as the backbone. A series of CT images were fed into COVIDNet to generate feature maps following the feature fusion layer. The feature fusion layer consists of 2 convolution layers. The final extracted features were fed into a dense layer and SoftMax activation to generate the prediction for COVID-19 pneumonia
More specifically, given that a CT study consists of a series of CT slices, we first preprocessed them and extracted the lung regions using U-net segmentation which was trained on kaggle dataset (https://www.kaggle.com/kmader/finding-lungs-in-ct-data). We augmented the training set with a random horizontal flip, random rotation, random scale, random translation, and random elastic transformation. The main code is available at https://github.com/LittleRedHat/COVID-19.
Performance in the training group was calculated as the mean value in five random groupings. Patients in the RWD group were used to testify the performance of our DL model.
Radiologist evaluation
In order to compare the performance of our AI model with the top human radiology experts, three senior experienced radiologists who were blinded to RT-PCR results were recruited and reviewed all de-identified chest CT images in the RWD group and scored each suspected case as COVID-19 or non-COVID-19 viral pneumonia. Information about the radiologists, including years in practice, average review time per case, cardiothoracic imaging fellowship, and COVID-19-specific training experience, is shown in Table S1.
Statistical analysis
All statistical analyses were performed with PyCharm IDE (version 3.5; JetBrains). The Shapiro-Wilk test was used to evaluate the distribution type and Bartlett’s test was used to evaluate the homogeneity of variance. Normally distributed data were expressed as mean ± standard deviation. Non-normally distributed data and ordinal data were expressed as median (1/4–1/3 quartile range). Categorical variables were summarized as counts and percentages. Comparisons of quantitative data were evaluated using the Mann-Whitney U test and Wilcoxon test. Comparisons of categorized data were evaluated by the chi-square test and Fisher test. A p value of < 0.05 was defined as with statistical significance. Missing data were omitted. The sensitivity and specificity for COVID-19 detection were calculated. The receiver operating characteristic (ROC) curve was plotted and the area under the curve (AUC) was calculated with the 95% confidence intervals (CIs) based on DeLong’s method.
Results
Study population characteristics
A total of 881 CT images from 696 patients, which were suspected to be COVID-19 pneumonia by radiologists, were included in our study to differentiate COVID-19 pneumonia patients from other non-COVID pneumonia patients. Among these patients, 470 were confirmed as COVID-19 and 226 were excluded by twice negative RT-PCR results.
The distribution of COVID-19 and non-COVID-19 patients in model-training and RWD groups was different. Of 380 patients, 227 were COVID-19 confirmed in the model-training group and 243/316 were proved to be infected by COVID-19 in the RWD group. Despite the differences between distributions, the COVID-19 patients had lower white blood cell count in both groups. Other clinical features did not show a significant difference between these 2 groups. Detailed information is summarized in Tables 1 and 2.
Model performance
Internal validation
The internal validation set composed of a total of 728 slices from 40 COVID-19 and 21 non-COVID-19 patients achieved the best performance. When the threshold was set at 0.685, our DL model achieved the best performance to differentiate COVID-19 from non-COVID-19 pneumonia with a sensitivity of 0.836, a specificity of 0.800, and an AUC of 0.906 (95% CI: 0.886–0.913) (Table 3, Fig. 3).
Real-world dataset
To validate our DL model’s general applicability in China, we obtained CT images from two institutions representing epidemic and non-epidemic areas of China. Our DL diagnostic model achieved 0.811 sensitivity, 0.822 specificity, and an AUC of 0.868 (95% CI: 0.851–0.876) for COVID-19 pneumonia versus all other types of pneumonia and the accuracy of our DL model in differentiating COVID-19 from non-COVID-19 pneumonia was 81% (95% CI: 77%, 84%). These results confirmed the high performance, accuracy, and general applicability of our DL model within China in this prospective RWD cohort (Fig. 3).
A comparison of the diagnostic performance between three senior experienced radiologists and the AI system is listed in Table 4. Our results indicated that our model using IDANNet could be used to distinguish COVID-19 from non-COVID-19 viral pneumonia with a non-inferior accuracy compared with that of experienced radiologists (Fig. 4).
The comparison of the diagnostic performance of RWD between three senior experienced radiologists and the AI system. The AI model operated at 81.1% sensitivity and 82.2% specificity (shown as the star) using a decision threshold set on the model development dataset. The performances of 3 experienced radiologists were labelled in dots
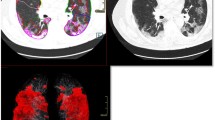
In order to show the interpretability of our model, we adopted the Grad-CAM to visualize the most important regions for making decision of the model. The attention heatmaps were fully generated by the model without additional manual annotation. Although features learned by DL models could reflect high-dimensional abstract mappings which were difficult for humans to sense but strongly associated with clinical outcomes, the attention regions were highly aligned with the ROIs acquired by human radiologists for diagnosis. Three typical cases are illustrated in Fig. 5.
Three attention heatmaps from the last “pooling” layer in our DL model. The attention regions were overlapping with the ROIs acquired by human radiologists. All these cases were diagnosed as possible COVID-19 pneumonia by radiologists but correctly distinguished out by the DL model. Thus, it is desirable to investigate what exact imaging features are DL model based on and how AI acquires the classification potential to improve the CT-based identification capability of clinicians and radiologists. A typical CT image in a COVID-19 pneumonia patient is illustrated in 1a–1c with subpleural GGO and “crazy paving” sign inside the lesion. A non-typical COVID-19 image is shown in 2a–2c with total consolidation in the right inferior lobe and a non-COVID viral pneumonia case is presented in 3a–3c with typical COVID-19 CT manifestations
Discussion
After the global outbreak of COVID-19, early screening and intervention of suspected COVID-19 patients including quarantine are necessary to guarantee the in-time treatment of infected patients and ensure other medical activities [2]. Chest CT scans serve as a screening method in clinically suspected patients currently. But since the radiological manifestation of COVID-19 lacks specificity, it is hard for radiologists to distinguish COVID-19 from other types of pneumonia. Furthermore, the diagnosis of COVID-19 was quite subjective and radiological diagnosis varied according to the incidence rate in the area. It was reported that in epidemic areas, the positive predictive value (PPV) of radiologists in differentiating COVID-19 from other types of pneumonia reached 65%, which we thought was partly due to the high incidence of COVID-19 and not barely based on the diagnosis ability of the radiologists [8]. When the epidemiological characteristics change, the PPV of radiological diagnosis tends to drop dramatically, and it is doubted whether chest CT is still valuable in such a situation. Therefore, we expected to develop an AI system which could help radiologists distinguish COVID-19 from other similar types of pneumonia in an objective way.
In this study, we designed a novel CNN-based DL model and the accuracy in internal validation data reached 90%. In order to diminish the risk of bias, enhance real-world clinical relevance, and improve reporting and transparency, a real-world cohort from 2 institutions in epidemic and non-epidemic areas was used to test the performance of our model. The AUC value in the RWD group was 86%, non-inferior to experienced radiologists, suggesting promising clinical usage with a higher evidence level.
Methodologically, AI-based segmentation is an important step for the quantification of COVID-19 images. The segmentation would help models focus on the features in regions of interests (ROIs) selected by humans. Different from other studies, we selected the suspected CT images in reference to the prior knowledge of radiologists and fed them to our DL model directly without any manual segmentation. Basically, the selection of suspected cases by radiologists was another type of “segmentation.” These two protocols mentioned above could be summarized as “segmentation first, diagnosis later” and “selection first, diagnosis later.” Except for the reason that the latter protocol could be directly applied to our clinical practice, there are two more reasons: First, none of the quantified parameters extracted from segmented regions was proved to be useful to disease diagnosis yet and most of them could not be clearly explained. Second, a robust segmentation network required a large number of ROIs for training and highly relied on the accuracy of ROIs drawn by humans, which was very costly and time-consuming while Dr. Zhang and his team have done a good job in this area [20]. After analyzing the prior selected images, our DL model could output diagnostic suggestions. Our testing result from RWD was non-inferior to the one from Zhang’s study and the sample size we used to train our model was much smaller. It would be interesting to compare the diagnostic efficiency between DL models trained by these 2 training protocols respectively in further studies.
In order to explain how our model worked, the important regions recognized by our model automatically were visualized by attention heatmaps. It could be observed that the suspicious pulmonary areas detected by our model were highly overlapping with the actual infected area recognized by radiologists. Some radiological features such as GGOs and crazy paving signs, which were reported to be crucial for COVID-19 diagnosis, were also included in the highlighted area labeled by the DL model, indicating that the high-dimensional features excavated by the DL model may reflect some radiological characteristics perceived by radiologists and make the quantification of these features possible.
Based on the prior evaluation of radiologists, our new DL model has the potential to be added to the clinical routine directly. When one suspected case was detected, radiologists could send the images directly to the DL model and obtain a diagnostic suggestion with an accuracy of over 80% which was convenient and feasible. Despite the good performance of our novel DL system, there are still several limitations. Firstly, we used RT-PCR results as the golden standard which was challenged frequently by its low positive rate. The sensitivity of chest CT to COVID-19 might be overestimated while the specificity would be underestimated. Secondly, the prognostic events, such as death or deterioration, were not taken into consideration in our study. Thirdly, we have not enrolled special population such as children and pregnant women.
Our established DL model was able to achieve accurate identification of COVID-19 from other suspected ones in the real-world situation on chest CT using prospective validation, which could aid in improving the clinical decision-making process. Future studies could be carried out to investigate a complete set of standard AI-based workflows for this global disaster from development to verification to integrate limited data resources and iterate existing AI products.
Abbreviations
- AUC:
-
Area under the curve
- CI:
-
Confidence interval
- CNN:
-
Convolutional neural network
- COVID-19:
-
Coronavirus disease 2019
- DL:
-
Deep learning
- GGO:
-
Ground-glass opacity
- IDANNet:
-
Identification and Analysis of New covid-19 Net
- ROC:
-
Receiver operating characteristic
- RT-PCR:
-
Reverse transcriptase-polymerase chain reaction
- RWD:
-
Real-world dataset
References
World Health Organization. Clinical management of severe acute respiratory infection (SARI) when COVID-19 disease is suspected, Interim guideline. Available via https://apps.who.int/iris/handle/10665/331446. Accessed 10 May 2020.
Guan WJ, Ni ZY, Hu Y et al (2020) Clinical characteristics of coronavirus disease 2019 in China. N Engl J Med 382(18):1708–1720
Wu Z, Jennifer M (2020) Characteristics of and important lessons from the coronavirus disease 2019 (COVID-19) outbreak in China: summary of a report of 72314 cases from the Chinese Center for Disease Control and Prevention. JAMA. https://doi.org/10.1001/jama.2020.2648
Wang W, Tang J, Wei F (2020) Updated understanding of the outbreak of 2019 novel coronavirus (2019-nCoV) in Wuhan, China. J Med Virol 92(4):441–447
Chen Y, Liu Q, Guo D (2020) Emerging coronaviruses: genome structure, replication, and pathogenesis. J Med Virol 92(4):418–423
Xie M, Chen Q (2020) Insight into 2019 novel coronavirus - an updated interim review and lessons from SARS-CoV and MERS-CoV. Int J Infect Dis 94:119–124
Fang Y, Zhang H, Xie J et al (2020) Sensitivity of chest CT for COVID-19: comparison to RT-PCR. Radiology:200432
Ai T, Yang Z, Hou H et al (2020) Correlation of chest CT and RT-PCR testing in coronavirus disease 2019 (COVID-19) in China: a report of 1014 cases. Radiology:200642
Xie X, Zhong Z, Zhao W, Zheng C, Wang F, Liu J (2020) Chest CT for typical 2019-nCoV pneumonia: relationship to negative RT-PCR testing. Radiology:200343
Shi H, Han X, Jiang N et al (2020) Radiological findings from 81 patients with COVID-19 pneumonia in Wuhan, China: a descriptive study. Lancet Infect Dis 20(4):425–434
Pan F, Ye T, Sun P et al (2020) Time course of lung changes on chest CT during recovery from 2019 novel coronavirus (COVID-19) pneumonia. Radiology 295(3):715–721
Rubin GD, Ryerson CJ, Haramati LB et al (2020) The role of chest imaging in patient management during the COVID-19 pandemic: a multinational consensus statement from the Fleischner Society. Chest. https://doi.org/10.1016/j.chest.2020.04.003
Kanne JP, Little BP, Chung JH, Elicker BM, Ketai LH (2020) Essentials for radiologists on COVID-19: an update - Radiology Scientific Expert Panel. Radiology:200527
Simpson S, Kay FU, Abbara S et al (2020) Radiological society of North America Expert Consensus statement on reporting chest CT findings related to COVID-19. Endorsed by the Society of Thoracic Radiology, the American College of Radiology, and RSNA. J Thorac Imaging. https://doi.org/10.1097/RTI.0000000000000524
Garcia-Ordas MT, Benitez-Andrades JA, Garcia-Rodriguez I, Benavides C, Alaiz-Moreton H (2020) Detecting respiratory pathologies using convolutional neural networks and variational autoencoders for unbalancing data. Sensors (Basel) 20(4):1214
Ning Z, Luo J, Li Y et al (2019) Pattern classification for gastrointestinal stromal tumors by integration of radiomics and deep convolutional features. IEEE J Biomed Heal Inform 23(3):1181–1191
Park B, Park H, Lee SM, Seo JB, Kim N (2019) Lung segmentation on HRCT and volumetric CT for diffuse interstitial lung disease using deep convolutional neural networks. J Digit Imaging 32(6):1019–1026
Gulshan V, Peng L, Coram M et al (2016) Development and validation of a deep learning algorithm for detection of diabetic retinopathy in retinal fundus photographs. JAMA 316(22):2402–2410
Kumar Shukla P, Kumar Shukla P, Sharma P et al (2020) Efficient prediction of drug-drug interaction using deep learning models. IET Syst Biol 14(4):211–216
Zhang K, Liu X, Shen J et al (2020) Clinically applicable AI system for accurate diagnosis, quantitative measurements and prognosis of COVID-19 pneumonia using computed tomography. Cell 181(6):1423–1433
Li L, Qin L, Xu Z et al (2020) Artificial intelligence distinguishes COVID-19 from community acquired pneumonia on chest CT. Radiology:200905
Singh D, Kumar V, Vaishali, Kaur M (2020) Classification of COVID-19 patients from chest CT images using multi-objective differential evolution-based convolutional neural networks. Eur J Clin Microbiol Infect Dis 39(7):1379–1389
Nagendran M, Chen Y (2020) Lovejoy CA, et al Artificial intelligence versus clinicians: systematic review of design, reporting standards, and claims of deep learning studies. BMJ 368:m689
Sherman RE, Anderson SA, Pan GJD et al (2016) Real-world evidence - what is it and what can it tell us? N Engl J Med 375(23):2293–2297
Lusignan SD, Crawford L, Munro N (2015) Creating and using real-world evidence to answer questions about clinical effectiveness. J Innov Heal Inform 22(3):368–373
Acknowledgments
We gratefully acknowledge the guidance from Dr. Daizong Ding and Dr. Jin Fan on data curation and extend our thanks to Miss Peng for all her kindness and support.
Funding
This project was supported by Shanghai Natural Science Foundation (Grant No. 18ZR1405700), Youth Program of the National Natural Science Foundation of China (Fund No. 81901697), Shanghai Sailing Program (Grant No. 18YF1403000), Shanghai Science and Technology Development (Fund No. 19511121204), Youth Medical Talents - Medical Imaging Practitioner Program (No. SHWRS(2020)_087), Shanghai Municipal Science and Technology Major Project (No. 2018SHZDZX01), and ZJ Lab.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Dr. Yin Bo.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors, Dr. Lu, has significant statistical expertise. No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was waived by the Institutional Review Board.
Ethical approval
Institutional Review Board approval was obtained.
Methodology
• prospective
• diagnostic study
• multi-center study
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 180 kb)
Rights and permissions
About this article
Cite this article
Xie, Q., Lu, Y., Xie, X. et al. The usage of deep neural network improves distinguishing COVID-19 from other suspected viral pneumonia by clinicians on chest CT: a real-world study. Eur Radiol 31, 3864–3873 (2021). https://doi.org/10.1007/s00330-020-07553-7
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07553-7