Abstract
Objectives
The goal of the present study was to classify the most common types of plain radiographs using a neural network and to validate the network’s performance on internal and external data. Such a network could help improve various radiological workflows.
Methods
All radiographs from the year 2017 (n = 71,274) acquired at our institution were retrieved from the PACS. The 30 largest categories (n = 58,219, 81.7% of all radiographs performed in 2017) were used to develop and validate a neural network (MobileNet v1.0) using transfer learning. Image categories were extracted from DICOM metadata (study and image description) and mapped to the WHO manual of diagnostic imaging. As an independent, external validation set, we used images from other institutions that had been stored in our PACS (n = 5324).
Results
In the internal validation, the overall accuracy of the model was 90.3% (95%CI: 89.2–91.3%), whereas, for the external validation set, the overall accuracy was 94.0% (95%CI: 93.3–94.6%).
Conclusions
Using data from one single institution, we were able to classify the most common categories of radiographs with a neural network. The network showed good generalizability on the external validation set and could be used to automatically organize a PACS, preselect radiographs so that they can be routed to more specialized networks for abnormality detection or help with other parts of the radiological workflow (e.g., automated hanging protocols; check if ordered image and performed image are the same). The final AI algorithm is publicly available for evaluation and extension.
Key Points
• Data from one single institution can be used to train a neural network for the correct detection of the 30 most common categories of plain radiographs.
• The trained model achieved a high accuracy for the majority of categories and showed good generalizability to images from other institutions.
• The neural network is made publicly available and can be used to automatically organize a PACS or to preselect radiographs so that they can be routed to more specialized neural networks for abnormality detection.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Machine learning is predicted to have a huge impact on the field of radiology [1], augmenting and assisting radiologists [2]. With new papers being published every week, one central question remains: What can machine learning do for the average radiologist? Currently, the majority of research in radiology seems to be focused on applying machine learning to the parts of the imaging pipeline that involve perception and reasoning (e.g., detection, quantification, and diagnostic reasoning) [3, 4]. However, due to various barriers (e.g., ethical, economical, and legal), this approach, while promising, may not be the optimal starting point for introducing artificial intelligence into the radiological workflow. Instead, artificial intelligence could be used as a tool for quality assurance and help with automating simple but tedious task encountered in clinical routine [5]. For example, one common challenge in a picture archiving and communication system (PACS) is that images are often labeled incorrectly in the corresponding DICOM tag. The problem of unreliable DICOM information was first demonstrated by Güld et al who found that the DICOM tag Body Part Examined was incorrect in 15.3% of cases [6]. This is not only problematic for the retrieval of images for the purpose of creating datasets but also hinders the development of imaging pipelines in which images are automatically routed to specific classification algorithms. Besides, many other parts of the radiological workflow rely on correctly labeled images. Thus, a neural network that can correctly classify and tag images could be used to check that exams are not repeated unnecessarily, control that the acquired image is the same as the one that was ordered, and streamline hanging protocols for optimal reporting on images. Because plain radiographs are still the most common type of imaging performed, a network for the classification of plain radiographs can have a meaningful impact on the radiological workflow. Therefore, the main goal of our study was to develop and validate a convolutional network to classify the most common types of plain radiographs (e.g., thorax pa, abdomen lateral). The final model will be made publicly available so that it can be evaluated and integrated into the radiological workflow.
Materials and methods
Radiographs
All radiographs from the year 2017 (N = 71,274) performed at our institution were retrieved from the PACS and categorized into 102 categories based on their DICOM metadata (study, series, and image description) according to the WHO manual of diagnostic imaging [7]. Because some categories contained only a small number of images, we limited ourselves to the 30 largest categories (n = 58,219), which accounted for 81.7% (58,219/71274) of all radiographs performed in the year 2017 at our institution. For these 30 categories, all images were reviewed again by one radiologist and misclassifications were corrected (i.e., discrepancies between DICOM information and actual image content). Table 1 shows the final dataset with all categories selected for the study and the number of images per class. For each of the 30 categories, 100 randomly selected images were set aside for internal validation (n = 3000) and the rest of the images was used as the training set (n = 55,219). To assess the generalizability of the results, we used images from other institutions, acquired with machines from multiple vendors, stored in our PACS (n = 5324) as an external validation set. To ensure that these images were labeled correctly, these images were manually labeled by two experienced radiologists because DICOM information could not be automatically processed, was missing, or was in several different languages. These images were not part of the training set and only used to validate the trained network. Table 1 shows the number of images per category for the external validation set.
Neural network training
All images were exported from the PACS as JPEG (Joint Photographic Expert Group) images and anonymized in the process. Using the images in the training set (n = 58,219), a pretrained MobileNet (Version 1.0) was retrained using oversampling—to account for imbalanced classes—with 22,000 training steps and a learning rate of 0.1. No image augmentation techniques were used. The network was trained on a standard MacBook Pro (Retina, 15–in., Late 2013, 16-GB DDR RAM, 2.3-GHz Quad-Core Intel Core i7).
Statistical analysis
Performance metrics, such as sensitivity, specificity, positive predictive value, and negative predictive value, were calculated using SPSS Version 26.0 [8].
Results
Internal validation
In the internal validation, the overall accuracy of the model in the validation set was 90.3% (95%CI: 89.2–91.3%). Because in this validation set the number of images in each class was equal (n = 100), the average sensitivity was the same as the accuracy (90.3%), indicating that, on average, 90.3% of images in each category were correctly classified by the model (see Table 2 for performance metrics for each individual class). As Table 2 shows, the distribution of the sensitivity of the model was rather balanced across categories, ranging between 61.0 and 100.0%. Eighteen out of 30 categories (60.0%) reached a sensitivity of over 90.0%, and 27 out of 30 categories (90.0%) reached a sensitivity of over 80.0%. Only the categories ankle lateral (sensitivity: 79%), lumbar spine lateral (sensitivity: 77%), and shoulder outlet (sensitivity: 61%) reached a sensitivity below 80.0%.
As for the other performance metrics, the model achieved an average specificity of 99.7%, indicating that, on average, 99.7% of images that were not part of a class were correctly labeled as not belonging to that class. The model achieved an average positive predictive value of 90.8%, indicating that out of all images predicted to belong to a certain class 90.8% of images did actually belong to that class. The average negative predictive value of the model was 99.7%.
External validation
In the external validation, the overall accuracy of the model in the unseen validation set was 94.0% (95%CI: 93.3–94.6%). The average sensitivity of the model was 93.2%, indicating that 93.2% of images in each category were correctly classified by the model (see Table 3 for performance metrics for each individual class). The sensitivity ranged between 75.0 and 100.0%. Twenty-three out of 30 categories (76.7%) reached a sensitivity of over 90.0%, and 29 out of 30 categories (96.7%) reached a sensitivity of over 80.0%. Only the category finger lateral (75%) scored below 80.0%.
As for the other performance metrics, the model achieved an average specificity of 99.8%, indicating that, on average, 99.8% of images that were not part of a class were correctly labeled as not belonging to that class. The model achieved an average positive predictive value of 88.6%, indicating that out of all images predicted to belong to a certain class 88.6% of images did actually belong to that class. The average negative predictive value of the model was 99.8%.
Discussion
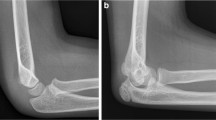
The goal of the present study was to create a neural network for practical applications in the imaging pipeline, e.g., to detect and correct errors in DICOM metadata, to rout radiographs to more specialized networks for abnormality detection, to check that exams are not repeated unnecessarily, to control that the acquired image is the same as the one that was ordered, and to streamline hanging protocols for optimal reporting on images. Our trained model was able to correctly classify the most common types of plain radiographs (e.g., thorax pa, abdomen lateral) and showed good generalizability in the internal (average accuracy: 90.3%) and external validation (average accuracy: 94.0%). However, an overall high accuracy does not necessarily mean that a model will be useful under real-world conditions. One important factor is a comparable level of high performance across all different categories. Combining the results from the internal and external validation set, performance across categories was generally balanced, with only four categories, ankle lateral (79.0%), lumbar spine lateral (77.0%), finger lateral (75.0%), and shoulder outlet (sensitivity: 61.0%) scoring below 80.0%. Taking a closer look at the errors in these categories revealed that the model tended to suggest similar categories and that the correct classification was in many cases the model’s second prediction (see Fig. 1). This may in part be due to suboptimal positioning in some images, for example, where the patient’s pain may have limited the radiographer’s ability to achieve perfect positioning. In contrast, highly standardized and unambiguous image categories (e.g., abdomen left lateral decubitus, patella axial, and chest pa/ap) showed perfect classification results with accuracies of up to 100.0%.
Examples of four images that were misclassified by the neural network. Images a and b actually belong to the class ankle lateral but were misclassified as ankle AP by the model. Images c and d actually belong to the class shoulder outlet but were misclassified as shoulder AP. The corresponding prediction values reflect the probability that the image belongs to a certain class, ranging from 0 to 100%. Higher values reflect a higher probability that an image belongs to a certain class
To further assess the performance of our model, it is important to compare its performance with other approaches to classify plain radiographs. Using a CNN and Radon transformation, Khatami et al achieved an accuracy of 90.3% for the validation set of the ImageCLEF2009 medical annotation task. This compares favorably with our own accuracy of 90.3% in the internal validation. However, it is difficult to compare performance on different datasets. To allow further assessment of our model, we will make it available so that other institutions are able to test the performance of the model using their own data.
Our study has some limitations: First, even though the 30 categories included in our study accounted for 81.7% (58,219/71274) of all radiographs performed in 1 year, an ideal system should also include the remaining 72 categories.
Second, the overall accuracy was only 90.3% so that every 1 in 10 images would still need some form of human intervention to be correctly classified. There are several reasons for this: (a) Current approaches are relatively “data-hungry,” which means they need large amounts of images to achieve a high accuracy. Until new techniques emerge that can produce better results with less data, the only option is for multiple institutions to pool their data for less frequent categories to achieve better performance for rare categories. (b) Performance was generally worse for suboptimal images. As mentioned before, performance of the network will depend on the number of low-quality images in the dataset, as high-quality images with little variation are classified more accurately. Because we did test the network on randomly sampled images from our real PACS, the accuracy of our model may be a more accurate predictor of real-world performance than testing the model on a curated data set with only few low-quality images.
With regard to the accuracy achieved in our study, it is important to note, however, that the errors of the model were not random as the model was particularly prone to mistaking similar categories and the correct option was usually among the top suggestions of the model. Furthermore, it would be feasible to use the probability values generated by the model to flag potentially incorrect predictions because we did find that the probability values for incorrect predictions were significantly lower (M = 68.2%, SD = 21.0%) compared with the probability values for the correct predictions (M = 95.2%, SD = 10.9%) (t(2708) = 35.7, p < .001, d = 1.61).
Taking into consideration the limitations of our model, the following applications for our AI algorithm are feasible: First, the model can be used to classify images and add or correct DICOM metadata. Even though human review is still needed, the workload can be significantly reduced. Considering that very common categories, such as chest pa/ap or chest lateral, were classified with a relatively high accuracy, large parts of a PACS can be corrected with little error. For instance, in our sample, chest imaging accounted for around 30.4% of radiographs performed in 1 year (21,697/71274). With the categories chest pa/ap and chest lateral achieving an accuracy of 100.0% in the internal validation, 30.4% of images in our sample could have been easily labeled using the AI algorithm. Furthermore, being a relatively low-stakes task compared with the detection of abnormalities, it would be relatively safe to deploy the model.
Second, as part of an automated imaging pipeline, the model can be used to route images to more specialized networks for abnormality detection. For instance, the model can first identify a chest image so that it can then be analyzed by a network specialized for detecting anomalies in chest radiographs [9], abdominal radiographs [10], or musculoskeletal radiographs [11,12,13,14]. Again, our model did not achieve perfect accuracy for all classes. However, we think that this does not rule out the deployment of the model. One possible solution for this problem would be to use both the average accuracy of a category as well as individual prediction values to decide how to process images. If an image is from a category with high accuracy (e.g., chest pa/ap) and the prediction value for that particular image is high (> 90.0%), it could be sent straight to a secondary network for abnormality detection. If an image is from a category with low accuracy (e.g., shoulder outlet) and the prediction value for that particular image is also low (< 70.0%), it could be flagged for human review.
In summary, we show that it is possible for a single institution to train a neural network to classify the most common categories of plain radiographs, which can then be used to clean up DICOM metadata or as part of an automated imaging pipeline. To encourage independent review and validation as well as to promote the introduction of new tools that may help radiologists and technicians with routine tasks, the final model will be made publicly available on GitHub (https://github.com/healthcAIr/NNCPR).
Abbreviations
- AI:
-
Artificial intelligence
- DICOM:
-
Digital Imaging and Communications in Medicine
- JPEG:
-
Joint Photographic Expert Group
- PACS:
-
Picture archiving and communication system
References
Choy G, Khalilzadeh O, Michalski M et al (2018) Current applications and future impact of machine learning in radiology. Radiology 288:318–328. https://doi.org/10.1148/radiol.2018171820
Langlotz CP (2019) Will artificial intelligence replace radiologists? Radiology Artificial Intelligence 1:e190058. https://doi.org/10.1148/ryai.2019190058
Hosny A, Parmar C, Quackenbush J, Schwartz LH, Aerts HJWL (2018) Artificial intelligence in radiology. Nat Rev Cancer 18:500–510. https://doi.org/10.1038/s41568-018-0016-5
Dratsch T, Caldeira L, Maintz D, Pinto dos Santos D (2020) Artificial intelligence abstracts from the European Congress of Radiology: analysis of topics and compliance with the STARD for abstracts checklist. Insights Imaging 11. https://doi.org/10.1186/s13244-020-00866-7
Harvey H (2018) Why AI will not replace radiologists. https://towardsdatascience.com/why-ai-will-not-replace-radiologists-c7736f2c7d80. Accessed 5 Sept 2020
Gueld MO, Kohnen M, Keysers D et al (2002) Quality of DICOM header information for image categorization. In: Siegel EL, Huang HK (eds) Medical imaging 2002: PACS and integrated medical information systems: design and evaluation, pp 280–287
Sandström S, Ostensen H, Pettersson H, Akerman K (2003) The WHO manual of diagnostic imaging. World Health Organisation
IBM Corp. (2019) IBM SPSS statistics for Macintosh
Rajpurkar P, Irvin J, Zhu K et al (2017) CheXNet: radiologist-level pneumonia detection on chest X-rays with deep learning. arXiv preprint arXiv:1711.05225
Cheng PM, Tejura TK, Tran KN, Whang G (2018) Detection of high-grade small bowel obstruction on conventional radiography with convolutional neural networks. Abdom Radiol 43:1120–1127. https://doi.org/10.1007/s00261-017-1294-1
Kim DH, MacKinnon T (2018) Artificial intelligence in fracture detection: transfer learning from deep convolutional neural networks. Clin Radiol 73:439–445. https://doi.org/10.1016/j.crad.2017.11.015
Tiulpin A, Thevenot J, Rahtu E, Lehenkari P, Saarakkala S (2018) Automatic knee osteoarthritis diagnosis from plain radiographs: a deep learning-based approach. Sci Rep 8:1727. https://doi.org/10.1038/s41598-018-20132-7
Urakawa T, Tanaka Y, Goto S, Matsuzawa H, Watanabe K, Endo N (2019) Detecting intertrochanteric hip fractures with orthopedist-level accuracy using a deep convolutional neural network. Skeletal Radiol 48:239–244. https://doi.org/10.1007/s00256-018-3016-3
Üreten K, Erbay H, Maraş HH (2020) Detection of rheumatoid arthritis from hand radiographs using a convolutional neural network. Clin Rheumatol 39:969–974. https://doi.org/10.1007/s10067-019-04487-4
Funding
Open Access funding provided by Projekt DEAL.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Prof. Dr. David Maintz.
Conflict of interest
The authors of this manuscript declare no relationships with any companies whose products or services may be related to the subject matter of the article.
Statistics and biometry
No complex statistical methods were necessary for this paper.
Informed consent
Written informed consent was given by all patients before image acquisition at the University Hospital Cologne.
Ethical approval
The need for institutional Review board approval was waived since only annoymized retrospective data was used.
Methodology
• Training of a neural network using plain radiographs from the PACS
• retrospective
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Dratsch, T., Korenkov, M., Zopfs, D. et al. Practical applications of deep learning: classifying the most common categories of plain radiographs in a PACS using a neural network. Eur Radiol 31, 1812–1818 (2021). https://doi.org/10.1007/s00330-020-07241-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-07241-6