Abstract
Objectives
To assess the diagnostic accuracy of machine learning (ML) in predicting isocitrate dehydrogenase (IDH) mutations in patients with glioma and to identify potential covariates that could influence the diagnostic performance of ML.
Methods
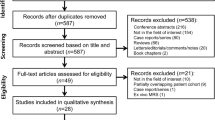
A systematic search of PubMed, Web of Science, and the Cochrane library up to 1 August 2019 was conducted to collect all the articles investigating the diagnostic performance of ML for prediction of IDH mutation in glioma. The search strategy combined synonyms for ‘machine learning’, ‘glioma’, and ‘IDH’. Pooled sensitivity, specificity, and their 95% confidence intervals (CIs) were calculated, and the area under the receiver operating characteristic curve (AUC) was obtained.
Results
Nine original articles assessing a total of 996 patients with glioma were included. Among these studies, five divided the participants into training and validation sets, while the remaining four studies only had a training set. The AUC of ML for predicting IDH mutation in the training and validation sets was 93% (95% CI 91–95%) and 89% (95% CI 86–92%), respectively. The pooled sensitivity and specificity were, respectively, 87% (95% CI 82–91%) and 88% (95% CI 83–92%) in the training set and 87% (95% CI 76–93%) and 90% (95% CI 72–97%) in the validation set. In subgroup analyses in the training set, the combined use of clinical and imaging features with ML yielded higher sensitivity (90% vs. 83%) and specificity (90% vs. 82%) than the use of imaging features alone. In addition, ML performed better for high-grade gliomas than for low-grade gliomas, and ML that used conventional MRI sequences demonstrated higher specificity for predicting IDH mutation than ML using conventional and advanced MRI sequences.
Conclusions
ML demonstrated an excellent diagnostic performance in predicting IDH mutation of glioma. Clinical information, MRI sequences, and glioma grade were the main factors influencing diagnostic specificity.
Key Points
• Machine learning demonstrated an excellent diagnostic performance for prediction of IDH mutation in glioma (the pooled sensitivity and specificity were 88% and 87%, respectively).
• Machine learning that used conventional MRI sequences demonstrated higher specificity in predicting IDH mutation than that based on conventional and advanced MRI sequences (89% vs. 85%).
• Integration of clinical and imaging features in machine learning yielded a higher sensitivity (90% vs. 83%) and specificity (90% vs. 82%) than that achieved by using imaging features alone.




Similar content being viewed by others
Abbreviations
- FA:
-
Fractional anisotropy
- HGG:
-
High-grade glioma
- HSROC:
-
Hierarchical summary receiver operating characteristic
- IDH:
-
Isocitrate dehydrogenase
- LGG:
-
Low-grade glioma
- MD:
-
Mean diffusion
- ML:
-
Machine learning
- PRISMA:
-
Preferred Reporting Items for Systematic Reviews and Meta-analyses
References
Louis DN, Perry A, Reifenberger G et al (2016) The 2016 World Health Organization classification of tumors of the central nervous system: a summary. Acta Neuropathol 131:803–820
Miller JJ, Shih HA, Andronesi OC, Cahill DP (2017) Isocitrate dehydrogenase-mutant glioma: evolving clinical and therapeutic implications. Cancer 123:4535–4546
Parsons DW, Jones S, Zhang X et al (2008) An integrated genomic analysis of human glioblastoma multiforme. Science 321:1807–1812
Yan H, Parsons DW, Jin G et al (2009) IDH1 and IDH2 mutations in gliomas. N Engl J Med 360:765–773
Kickingereder P, Sahm F, Radbruch A et al (2015) IDH mutation status is associated with a distinct hypoxia/ angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep 5:16238
Xing Z, Yang X, She D, Lin Y, Zhang Y, Cao D (2017) Noninvasive assessment of IDH mutational status inWorld Health Organization grade II and III astrocytomas using DWI and DSC-PWI combined with conventionalMR imaging. AJNR Am J Neuroradiol 38:1138–1144
Suh CH, Kim HS, Jung SC, Choi CG, Kim SJ (2019) Imaging prediction of isocitrate dehydrogenase (IDH) mutation in patients with glioma: a systemic review and meta-analysis. Eur Radiol 29:745–758
Suh CH, Kim HS, Paik W et al (2019) Falsepositive measurement at 2-hydroxyglutarate MR spectroscopy in isocitrate dehydrogenase wild-type glioblastoma: a multifactorial analysis. Radiology 291(3):752–762
Pesapane F, Codari M, Sardanelli F (2018) Artificial intelligence in medical imaging: threat or opportunity? Radiologists again at the forefront of innovation in medicine. Eur Radiol Exp 2:35
Qi S, Yu L, Li H et al (2014) Isocitrate dehydrogenase mutation is associated with tumor location and magnetic resonance imaging characteristics in astrocytic neoplasms. Oncol Lett 7:1895–1902
Zhou H, Vallieres M, Bai HX et al (2017) MRI features predict survival and molecular markers in diffuse lowergrade gliomas. Neuro Oncol 19:862–870
Aerts HJ, Velazquez ER, Leijenaar RT et al (2014) Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun 5:4006
Liberati A, Altman DG, Tetzlaff J et al (2009) The PRISMA statement for reporting systematic reviews and meta-analyses of studies that evaluate health care interventions: explanation and elaboration. J Clin Epidemiol 62:e1–e34
Whiting PF, Rutjes AW, Westwood ME et al (2011) QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med 155:529–536
Zamora J, Abraira V, Muriel A, Khan K, Coomarasamy A (2006) Meta-DiSc: a software for meta-analysis of test accuracy data. BMC Med Res Methodol 6:31
Deville WL, Buntinx F, Bouter LM et al (2002) Conducting systematic reviews of diagnostic studies: didactic guidelines. BMC Med Res Methodol 2:9
Deeks JJ, Macaskill P, Irwig L (2005) The performance of tests of publication bias and other sample size effects in systematic reviews of diagnostic test accuracy was assessed. J Clin Epidemiol 58:882–893
Zhang X, Tian Q, Wang L et al (2018) Radiomics strategy for molecular subtype stratification of lower-grade Glioma: detecting IDH and TP53 mutations based on multimodal MRI. J Magn Reson Imaging 48:916–926
Lohmann P, Lerche C, Bauer EK et al (2018) Predicting IDH genotype in gliomas using FET PET radiomics. Sci Rep 8:13328
Liang S, Zhang RG, Liang DY et al (2018) Multimodal 3D DenseNet for IDH genotype prediction in gliomas. Genes (Basel) 9(8)
Li ZC, Bai H, Sun Q et al (2018) Multiregional radiomics profiling from multiparametric MRI: identifying an imaging predictor of IDH1 mutation status in glioblastoma. Cancer Med 7:5999–6009
De Looze C, Beausang A, Cryan J et al (2018) Machine learning: a useful radiological adjunct in determination of a newly diagnosed glioma’s grade and IDH status. J Neurooncol 139:491–499
Chen L, Zhang H, Lu J et al (2018) Multilabel nonlinear matrix completion with transductive multi-task feature selection for jointMGMTand IDH1 status prediction of patient with high-grade gliomas. IEEE Trans Med Imaging 37:1775–1787
Yu J, Shi Z, Lian Y et al (2017) Noninvasive IDH1 mutation estimation based on a quantitative radiomics approach for grade II glioma. Eur Radiol 27:3509–3522
Choi KS, Choi SH, Jeong B (2019) Prediction of IDH genotype in gliomas with dynamic susceptibility contrast perfusion MR imaging using an explainable recurrent neural network. Neuro Oncol. https://doi.org/10.1093/neuonc/noz095
Tan Y, Zhang ST, Wei JW et al (2019) A radiomics nomogram may improve the prediction of IDH genotype for astrocytoma before surgery. Eur Radiol 29:3325–3337
Zhang BQ, Chang K, Ramkissoon S et al (2017) Multimodal MRI features predict isocitrate dehydrogenase genotype in high-grade gliomas. Neuro Oncol 19:109–117
Wijnenga MMJ, French PJ, Dubbink HJ et al (2018) The impact of surgery inmolecularly defined low-grade glioma: an integrated clinical, radiological, and molecular analysis. Neuro Oncol 20:103–112
Nakae S, Murayama K, Sasaki H et al (2017) Prediction of genetic subgroups in adult supra tentorial gliomas by pre- and intraoperative parameters. J Neurooncol 131:403–412
Shibahara I, Sonoda Y, Shoji T et al (2015) Malignant clinical features of anaplastic gliomas without IDH mutation. Neuro Oncol 17:136–144
Tietze A, Choi C, Mickey B et al (2018) Noninvasive assessment of isocitrate dehydrogenase mutation status in cerebral gliomas by magnetic resonance spectroscopy in a clinical setting. J Neurosurg 128:391–398
Funding
This study was supported by a grant from the Natural Science Foundation of Guangdong Province (2017A030313676).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Guarantor
The scientific guarantor of this publication is Jianping Chu.
Conflict of interest
The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article.
Statistics and biometry
One of the authors (Yingqian Huang) has significant statistical expertise (2 years of experience in a systematic review and meta-analysis).
Informed consent
Written informed consent was not required for this study because of the nature of our study, which was a systematic review and meta-analysis.
Ethical approval
Institutional Review Board approval was not required because of the nature of our study, which was a systematic review and meta-analysis.
Methodology
• A systematic review and meta-analysis performed at one institution
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
ESM 1
(DOCX 58 kb)
Rights and permissions
About this article
Cite this article
Zhao, J., Huang, Y., Song, Y. et al. Diagnostic accuracy and potential covariates for machine learning to identify IDH mutations in glioma patients: evidence from a meta-analysis. Eur Radiol 30, 4664–4674 (2020). https://doi.org/10.1007/s00330-020-06717-9
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-020-06717-9