Abstract
Objectives
Deep medullary veins support the venous drainage of the brain and may display abnormalities in the context of different cerebrovascular diseases. We present and evaluate a method to automatically detect and quantify deep medullary veins at 7 T.
Methods
Five participants were scanned twice, to assess the robustness and reproducibility of manual and automated vein detection. Additionally, the method was evaluated on 24 participants to demonstrate its application. Deep medullary veins were assessed within an automatically created region-of-interest around the lateral ventricles, defined such that all veins must intersect it. A combination of vesselness, tubular tracking, and hysteresis thresholding located individual veins, which were quantified by counting and computing (3-D) density maps.
Results
Visual assessment was time-consuming (2 h/scan), with an intra-/inter-observer agreement on absolute vein count of ICC = 0.76 and 0.60, respectively. The automated vein detection showed excellent inter-scan reproducibility before (ICC = 0.79) and after (ICC = 0.88) visually censoring false positives. It had a positive predictive value of 71.6 %.
Conclusion
Imaging at 7 T allows visualization and quantification of deep medullary veins. The presented method offers fast and reliable automated assessment of deep medullary veins.
Key Points
• Deep medullary veins support the venous drainage of the brain
• Abnormalities of these veins may indicate cerebrovascular disease and quantification is needed
• Automated methods can achieve this and support human observers
• The presented method provides robust and reproducible detection of veins
• Intuitive quantification is provided via count and venous density maps
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Deep medullary veins drain venous blood from the white matter towards the subependymal veins of the lateral ventricles [1, 2]. They are organized in a typical fan-pattern [3], which can be appreciated in Fig. 1. Abnormalities of the deep medullary veins may be involved in various forms of cerebral small vessel disease [4–8].
Transversal (left) and coronal (right) view of the second echo of a dual-echo gradient echo 7 T MRI sequence. In these minimum intensity projections of ten slices (resulting slab thickness; left: 3.9 mm, right: 3.5 mm), the deep medullary veins are clearly visible (encircled in one hemisphere). In the coronal view, the typical fan-pattern can be appreciated. The veins drain venous blood towards the subependymal veins, such as the caudate vein of Schlesinger (arrow)
With advanced imaging sequences and (ultra-)high field 7 T MRI, deep medullary veins can be visualized in vivo with high spatial resolution [9–11]. This would allow for further evaluation of the role of these veins and venous changes in cerebral small vessel disease, and in the context of ageing and cognitive decline. Since these veins are numerous, computer assisted detection and quantification may support a robust and reproducible assessment.
In this work, we propose and evaluate an automated method for the detection and quantification of deep medullary veins on 7 T MR images. We devise a region-of-interest (ROI) that captures the 3-D distribution of the veins. Individual veins are automatically detected and quantified based on number and density. Robustness and reproducibility of visual and automated vein detection are assessed; potential use-cases are demonstrated with a group of healthy senior participants.
Materials and methods
Study design
The proposed method was evaluated by assessing the inter-observer, intra-observer, and inter-scan reproducibility. Visual assessments of veins were compared with automated assessments, before and after censoring false positive detections. Two observers were involved and five participants were scanned twice, to assess inter-scan differences in visual and automated assessments. Details follow in the next sections.
Participants
A total of 35 participants was included, separated into two groups: Gval and Gapp. Written informed consent was given by all participants and the study was approved by the local institutional review board and ethical committee.
Gval was used to validate the method and assess its robustness and reproducibility. It included five healthy participants (2 male; age: 28 ± 4.7 years). These participants underwent MRI acquisition twice on the same day, with repositioning in between.
Gapp was formed by 30 aged individuals (17 male; 70 ± 3.8 years) included from a previous study [12], to demonstrate applications of the method. Participants with contraindications for 7 T MRI were excluded. After MRI acquisition, six participants were excluded because they had ungradable images owing to motion artefacts, leaving 24 participants.
MRI
MRI acquisition was performed on a 7 T whole-body system (Philips Healthcare, Cleveland, OH, USA). The protocol included a 3-D dual-echo gradient echo sequence (TR/TE1/TE2: 20/6.9/15.8 ms, acquisition matrix: 400 × 400 × 172, acquired voxelsize: 0.5 × 0.5 × 0.7 mm3, reconstructed voxelsize: 0.39 × 0.39 × 0.35 mm3, flip angle: 12°, bandwidth: 203 Hz/pixel, flow compensated gradients, sensitivity encoding factor: 2.5 (right-left), scan duration: 9:18) and a 3D T1-weighted sequence (TR/TI/TE: 4.8/1240/2.2 ms, reconstructed voxelsize: 0.66 × 0.66 × 0.50 mm3) [12].
Detection of deep medullary veins
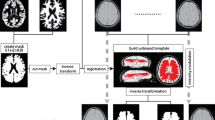
Automated detection of deep medullary veins consists of two steps: definition of an ROI in which the veins are located and the detection of individual veins. For the ROI, we have proposed to use an expanded ventricular surface [13], which the deep medullary veins must intersect to reach the subependymal veins (see Fig. 2-left). This surface is defined at 5 mm from the ventricles, restricted inferiorly by a plane that touched the genu and splenium of the corpus callosum and medially a region of 1.5 cm around the interhemispheric fissure was removed. A combination of the vesselness filter [14, 15], tubular tracking [16], and hysteresis thresholding located individual veins. Full details are given in the supplementary materials.
A minimum intensity projection slab of 3.5 mm (ten slices), showing that the deep medullary veins draining the deep white matter must intersect with the expanded ventricular surface (in red, ventricle segmentation in green) to reach the subependymal veins to which they connect. Right: the veins (white dots) that are detected at the location where they intersect with the ROI
Quantification
Veins were quantified by counting and computing venous density. Venous density was defined as the count divided by the area of the expanded ventricular surface.
Venous density maps were computed to intuitively visualize the distribution of veins. To be able to compare density maps between subjects or groups, all detected veins were transformed to the MNI152 template [17–19]. For each point on the expanded ventricular surface in MNI152 template space, the number of veins within a 15 mm radius was counted. An example result is shown in Fig. 3.
A 3-D rendering of the venous density map of all 24 participants in Gapp combined. This map was generated by transforming all detected veins from the scans of the participants to the MNI152 template. For each point on the expanded ventricular surface, all veins within a radius of 15 mm are counted. The colours represent a vein count ranging from low (blue, at least one vein at that location in a participant of Gapp) to high (red, on average four veins within a 15 mm radius at that location)
To visualize the vein distribution in the deep white matter, a 3-D venous density map was created. All detected veins were used as seed points in a tubular tracking algorithm [16] to track each individual deep medullary vein (see Fig. 4). All resulting tracked vein points were transformed to the MNI152 template. For each individual voxel of the MNI152 template, we counted the number of tracked vein points within a 15 mm radius (see Fig. 5) [20].
Deep medullary veins (orange) as tracked by a 3-D tubular tracking algorithm. The vein-points detected on the expanded ventricular surface (see Fig. 2) were used as seedpoints, and tracking was performed into the deep white matter. Left: transversal minimum intensity projection showing the individual tracked vein points (small orange dots). Right: 3-D rendering of the deep medullary veins as orange tubes, the white surface is the expanded ventricular surface
Venous density map of all participants in Gapp, where all reconstructed veins are transformed to the MNI152 template. The MNI152 template is shown for a number of slices (Z) and the venous density is overlaid in colour (blue = low density, red = high density). Each reconstructed vein consists of many individual points (see Fig. 4-left), approximately one point per venous voxel. The given density denotes the number of vein points within a 15 mm radius of each MNI152 template voxel
Evaluation
Our proposed method was validated against a visual assessment of deep medullary veins. For this, we used the first scans of the participants in the validation group Gval. Two human observers performed visual assessments of these scans. Observer 1 annotated an ROI of 60 slices, to which the automated detections were compared. The top of this ROI was set five slices above the roof of the lateral ventricles. The inter-observer reproducibility was assessed on 30 consecutive slices that were rated by both observers. Observers annotated veins where they intersected the expanded ventricular surface. Observer 1 annotated twice, with two weeks in between the assessments to assess the intra-observer reproducibility.
Next, observer 1 censored the false positive detections of the automated vein detection. Censoring of the first scans of Gval was done twice, to assess the intra-observer reproducibility.
By using the second scan of the participants in the validation group Gval, the inter-scan robustness and reproducibility of the visual and automated vein detection were determined. Observer 1 annotated the second scan of the participants in Gval. In addition, the reproducibility of the automated ROI definition was assessed by comparing each individual step of the presented method between the repeated scans of Gval.
Finally, all scans in Gapp were censored by observer 1. The positive predictive value (PPV = true veins / (true veins + censored false positives)) was computed.
All statistics were computed with SPSS 20 (IBM Corp.) The intra-class correlation coefficient (ICC) was used as the measure for reproducibility, assessing both the absolute agreement (ICCA) and consistency (ICCC) between measurements. ICCA|C indicates both measurements (ICCA and ICCC, respectively). A high ICCA required observers to agree on the exact vein count. The ICCC required observers to agree on the relative ordering of participants (few/many veins), but not on the exact vein count.
Results
Visually annotating deep medullary veins required, on average, 2 hours per scan. The intra-observer reproducibility on vein count was moderate (ICCA|C = 0.67|0.94). The inter-observer reproducibility was low to moderate with an ICCA = 0.45 and 0.74 (average: 0.60), but had a very good consistency with ICCC = 0.87 and 0.94 (average: 0.91). Censoring the false positives after automated vein detection required ±15 minutes per scan. The intra-observer reproducibility of the censoring is excellent, with an ICCA|C = 0.98|0.98. All tests and ICC values are summarized in Table 1.
The inter-scan reproducibility (between both scans of Gval, assessed by observer 1) was moderate with an ICCA|C = 0.72|0.68. The discrepancies between the repeated scans of Gval occurred mostly for smaller veins that consist heavily of partial volume voxels. The inter-scan reproducibility of the automated vein count before censoring is good with an ICCA|C = 0.79|0.76. The inter-scan reproducibility after censoring was very good with an ICCA|C = 0.88|0.85. This is summarized in Table 2.
The reproducibility of the automated ROI definition was excellent, having an ICCA > 0.98 for all individual steps. Computations required about 25 minutes per participant, using one core of a standard workstation.
Quantification
The average deep medullary vein count per participant after censoring was (mean ± sd) 365 ± 83 (Gval: 319 ± 71; Gapp: 384 ± 81). The PPV of the automated vein detection is on average 71.6 % ± 10.4 % (Gval: 77.8 % ± 7.0 % for 5×2 scans; Gapp: 69.0 % ± 10.6 % for 24 scans).
Visual inspection does not reveal large differences in venous density between the repeated scans of the participants in Gval. An average venous density map of the 24 participants in Gapp is given in Fig. 3. A high venous density is visible around the frontal horns of the lateral ventricles, where many deep medullary veins drain to the caudate veins of Schlesinger. More superiorly, another high venous density can be seen around the subependymal veins to which the deep medullary veins drain. At this location, an asymmetry is visible where the right side of the brain seems to have a higher venous density (3.9 versus 3.3 in the left hemisphere, but this difference is not statistically significant).
All reconstructed veins for the participants in Gapp were transformed to the MNI152 template and that result can be seen in Fig. 5. Again, superiorly, a slight asymmetry is visible. Next to this, many veins are tracked towards the thalamus and the basal veins.
Discussion
The presented method offers reliable, 3-D assessment of deep medullary veins on 7 T brain MRI, with an excellent reproducibility and a good PPV. Compared to visual detection, it is much less laborious and not subject to high intra- and inter-observer variability. The method offers new MRI measures that have potential to study the involvement of deep medullary veins in various conditions, such as cerebrovascular disease or dementia.
Visual assessment proved to be difficult, reflected by its low to moderate reproducibility. This is likely caused by partial volume effects for small veins, where each observer has a (different) “intrinsic cut-off” when some hypointense structure is denoted as a vein or not. Furthermore, these small partial volume veins cannot be reliably visualized from one scan to another, leading to the discrepancies between the repeated scans of Gval. Therefore, it is essential to focus on prominent deep medullary veins that can be reliably detected. When comparing scans from participants or groups, a robust and reproducible detection is important. The automated detection showed robust and reproducible results, owing to its fixed and deterministic behaviour.
The scan quality is of high importance for a reliable detection of deep medullary veins, both visually and automatically. The scans of the (younger) participants in Gval were of high quality, but some of the (senior) participants in Gapp were excluded owing to poor scan quality. The senior participants proved to be less capable to refrain from accidental motion; which is not a limitation of the presented method per se, but a more general problem in acquiring high quality images of elderly participants or patients [21]. Applying the method on lower field strength (e.g., 1.5 T or 3 T) should be feasible, but might require adaptations of the parameters and dedicated acquisition sequences, such as in the work of Ge et al. and Yan et al. on 3 T images [6, 22].
A limitation of the method is that visual censoring was still needed to remove false positive detections. For the presented method to be implemented in a fully automated workflow, without any human observer interaction, additional image processing is needed to remove the false positive detections. However, the uncensored inter-scan reproducibility is sufficient for quantifications that do not require a high PPV. The PPV is mainly determined by the selected values for some thresholds in the method (see supplementary materials). The more faint veins were not detected by this automated method, because the vesselness response for faint veins does not exceed the detection threshold. Adapting the thresholds to detect these faint veins will result in a lower PPV. However, the visual assessment showed that these veins also cannot be reliably visualized on repeated acquisitions of the same participant. Therefore, the inability to detect such faint veins actually contributes to the robustness and reproducibility of the detection and quantification. Another limitation of our method is that the visibility of veins may depend on the oxygenation of venous blood, because a higher concentration of deoxygenated haemoglobin will increase the susceptibility effects of the blood. This should be considered when comparing number and density of veins between persons, especially if cerebral blood flow could be reduced, such as for example in dementia [23].
The applications of the presented method are diverse. The potential clinical relevance of deep medullary veins on MRI was recently pointed out by several studies. A reduced venous density in the white matter of patients with Cerebral Autosomal-Dominant Arteriopathy With Subcortical Infarcts and Leukoencephalopathy (CADASIL) compared with controls was reported by De Guio et al. [4], and increased numbers of voxels of deep medullary veins in white matter hyperintensities by Yan et al. [6]. In patients with acute stroke, Mucke et al. [24] reported that an asymmetric appearance of deep medullary veins on MRI predicted stroke severity. Furthermore, both Ge et al. and Sinnecker et al. reported less visible deep medullary veins in patients with multiple sclerosis (MS) [5, 22]. Compared with these studies, the presented method offers new quantification measures of deep medullary veins that can be used in future studies. Precise, 3-D vein counts and venous density maps can be compared within or between groups of participants. The tracking of veins back into the deep white matter allows for a full brain analysis of spatial differences in deep medullary veins, and the computation of measures such as length and tortuosity of individual veins. Comparing the venous density maps in MNI152 template space might reveal spatial differences in the distribution of veins between groups, which cannot be revealed solely by vein count. Because all veins are in the same MNI152 template space, voxel-wise comparisons can be made easily and tested for statistical significant differences between groups. Also, the 3-D segmentation offers the possibility to apply new techniques such as quantitative susceptibility mapping on veins [25]. Our method offers new possibilities to study the role of venous changes in brain diseases, for example cerebrovascular disease or MS, and their spatial relation with brain lesions.
Abbreviations
- ICC:
-
Intra-class correlation coefficient
- ICCA :
-
ICC on the absolute agreement between measurements
- ICCC :
-
ICC on the consistency between measurements
- ICCA|C :
-
Both ICCA and ICCC
- PPV:
-
Positive predictive value
References
Hooshmand I, Rosenbaum AE, Stein RL (1974) Radiographic anatomy of normal cerebral deep medullary veins: criteria for distinguishing them from their abnormal counterparts. Neuroradiology 7(2):75–84
Friedman DP (1997) Abnormalities of the deep medullary white matter veins: MR imaging findings. AJR Am J Roentgenol 168(4):1103–1108
Schlesinger B (1939) The venous drainage of the brain, with special reference to the Galenic system. Brain 62(3):274–291
De Guio F, Vignaud A, Ropele S et al (2014) Loss of venous integrity in cerebral small vessel disease A 7-T MRI study in Cerebral Autosomal-Dominant Arteriopathy With Subcortical Infarcts and Leukoencephalopathy (CADASIL). Stroke 45:2124–2126
Sinnecker T, Bozin I, Dörr J et al (2013) Periventricular venous density in multiple sclerosis is inversely associated with T2 lesion count: a 7 Tesla MRI study. Mult Scler 19(3):316–325
Yan S, Wan J, Zhang X et al (2014) Increased visibility of deep medullary veins in leukoaraiosis: a 3-T MRI study. Front Aging Neurosci. doi:10.3389/fnagi.2014.00144
Moody DM, Brown WR, Challa VR, Ghazi-Birry HS, Reboussin DM (1997) Cerebral microvascular alterations in aging, leukoaraiosis, and Alzheimer's disease. Ann N Y Acad Sci 826(1):103–116
Moody DM, Thore CR, Anstrom JA, Challa VR, Langefeld CD, Brown WR (2004) Quantification of afferent vessels shows reduced brain vascular density in subjects with leukoaraiosis. Radiology 233(3):883–890
Haacke EM, Xu Y, Cheng YCN, Reichenbach JR (2004) Susceptibility weighted imaging (SWI). Magn Reson Med 52(3):612–618
Zwanenburg JJM, Versluis MJ, Luijten PR, Petridou N (2011) Fast high resolution whole brain T2* weighted imaging using echo planar imaging at 7T. Neuroimage 56(4):1902–1907
van Veluw SJ, Zwanenburg JJM, Hendrikse J, van der Kolk AG, Luijten PR, Biessels GJ (2014) High resolution imaging of cerebral small vessel disease with 7 T MRI. In: Trends in neurovascular interventions. 125–130. doi:10.1007/978-3-319-02411-0_21
Brundel M, Reijmer YD, van Veluw SJ, et al. (2014) Cerebral microvascular lesions on High-Resolution 7T MRI in patients with type 2 diabetes. Diabetes 63(10):3523–3529
Kuijf HJ, Bouvy WH, Zwanenburg JJM, Viergever MA, Biessels GJ, Vincken KL (2015) Automated detection of periventricular veins on 7 T brain MRI. In: Proc. SPIE 9413, Medical Imaging 2015: Image Processing, 94132T. doi:10.1117/12.2080952
Frangi AF, Niessen WJ, Vincken KL, Viergever MA (1998) Multiscale vessel enhancement filtering. In: Medical Image Computing and Computer-Assisted Interventation—MICCAI’98. 130–137. doi:10.1007/BFb0056195
Sato Y, Nakajima S, Shiraga N et al (1998) Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images. Med Image Anal 2(2):143–168
Friman O, Hindennach M, Kühnel C, Peitgen HO (2010) Multiple hypothesis template tracking of small 3D vessel structures. Med Image Anal 14(2):160–171
Fonov VS, Evans AC, McKinstry RC, Almli CR, Collins DL (2009) Unbiased nonlinear average age-appropriate brain templates from birth to adulthood. Neuroimage 47:S102
Fonov V, Evans AC, Botteron K, Almli CR, McKinstry RC, Collins DL (2001) Unbiased average age-appropriate atlases for pediatric studies. Neuroimage 54(1):313–327
Klein S, Staring M, Murphy K, Viergever MA, Pluim JP (2010) Elastix: a toolbox for intensity-based medical image registration. IEEE Trans Med Imaging 29(1):196–205
Muja M, Lowe DG (2009) Fast approximate nearest neighbors with automatic algorithm configuration. VISAPP 1:331–340
Zeng LL, Wang D, Fox MD et al (2014) Neurobiological basis of head motion in brain imaging. Proc Natl Acad Sci U S A 111(16):6058–6062
Ge Y, Zohrabian VM, Osa EO et al (2009) Diminished visibility of cerebral venous vasculature in multiple sclerosis by susceptibility-weighted imaging at 3.0 Tesla. J Magn Reson Imaging 29(5):1194
Binnewijzend MAA, Kuijer JPA, Benedictus MR et al (2013) Cerebral blood flow measured with 3D pseudocontinuous arterial spin-labeling MR imaging in Alzheimer disease and mild cognitive impairment: a marker for disease severity. Radiology 267(1):221–230
Mucke J, Möhlenbruch M, Kickingereder P et al (2015) Asymmetry of deep medullary veins on susceptibility weighted MRI in patients with acute MCA stroke is associated with poor outcome. PLoS One 10(4):e0120801
Haacke EM, Tang J, Neelavalli J, Cheng YCN (2010) Susceptibility mapping as a means to visualize veins and quantify oxygen saturation. J Magn Reson Imaging 32(3):663–676
Acknowledgments
The scientific guarantor of this publication is Geert Jan Biessels. The authors of this manuscript declare no relationships with any companies, whose products or services may be related to the subject matter of the article. No complex statistical methods were necessary for this paper. Institutional Review Board approval was obtained. Written informed consent was obtained from all subjects (patients) in this study. Some study subjects or cohorts have been previously reported in Brundel M, Reijmer YD, van Veluw SJ, et al. Cerebral microvascular lesions on High-Resolution 7 T MRI in patients with type 2 diabetes. Diabetes. 2014;63(10):3523–3529. DOI: 10.2337/db14-0122.
Methodology: retrospective, observational, performed at one institution. This study was financially supported by the project Brainbox (Quantitative analysis of MR brain images for cerebrovascular disease management), funded by the Netherlands Organisation for Health Research and Development (ZonMw) with in kind matching by Philips, the University Medical Center Utrecht and Eindhoven University of Technology, in the framework of the research programme IMDI (Innovative Medical Devices Initiative); project 104002002. This work was also supported by a Vidi grant from ZonMw, The Netherlands Organisation for Health Research and Development [91711384], to GJB. JZ received funding from the European Research Council under the European Union’s Seventh Framework Programme (FP7/2007-2013) / ERC grant agreement № 337333. We acknowledge the use of MeVisLab (MeVis Medical Solutions AG, Bremen, Germany). The authors thank Zoë Dalmijn for her contributions to the validation study and data analysis.
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
ESM 1
(DOCX 24.9 kb)
Fig. S1
Left: original T1 image. Middle: CSF segmentation as obtained from SPM12b. Right: the ventricles are extracted from the CSF segmentation by registering with the MNI152 template atlas and thresholding at 80 %. The resulting binary mask is transformed to the second echo of the gradient echo scan. (JPG 823 kb)
Fig. S2
Top: a minimum intensity projection slab of 3.5 mm (ten slices), showing the resulting expanded ventricular surface (red) and the midsagittal plane (white). Note that the expanded ventricular surface is a 3-D surface that has been flattened over the slab of ten slices in this illustration. Bottom: sagittal slice through the interhemispheric fissure, showing the automatically positioned plane that touches the genu and splenium of the corpus callosum, defining the inferior side of the ROI. (JPG 944 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution-NonCommercial 4.0 International License (http://creativecommons.org/licenses/by-nc/4.0/), which permits any noncommercial use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made.
About this article
Cite this article
Kuijf, H.J., Bouvy, W.H., Zwanenburg, J.J.M. et al. Quantification of deep medullary veins at 7 T brain MRI. Eur Radiol 26, 3412–3418 (2016). https://doi.org/10.1007/s00330-016-4220-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00330-016-4220-y