Abstract
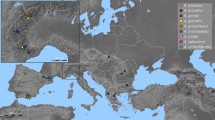
Unlike the Arctic flora, with many flowering plant species offering opportunities to study evolutionary processes, the Antarctic flora offers only two. One of them is the Antarctic grass Deschampsia antarctica E. Desv., whose distribution spans from northern Patagonia (ca. 38°S) down to Alamode Island (ca. 68°S), in the west side of the Antarctic Peninsula. While some aspects of Antarctic plants have been extensively studied (e.g., anatomy, physiology, genetics), little is known about the related Patagonian populations. Particularly in cytogenetics, no single study has focused on continental populations and its relationships with the Antarctic plants. The combination of traditional fluorescent in situ hybridization (FISH) with a phylogenetic framework highlights the importance of cytogenetics in plant evolutionary studies, by allowing comparison of chromosome characters in phylogenetically related individuals. Most used characters for this purpose are the chromosome number, karyotype morphology and patterns of repetitive DNA. These were used to compare distant populations of D. antarctica in a phylogenetic framework, to obtain a first view of the cytogenetic structure of the species along its distribution. Patagonian populations have greater variability in the chromosomal and molecular characters, while Antarctic populations are very alike, hinting at a South American origin hypothesis. A polyploid population is reported for the first time, located on Central Patagonia populations, close to the northern limit of distribution range. Cytogenetic characteristics suggest that hybridization processes could have played an important role in the evolution of the genome of D. antarctica.








Similar content being viewed by others
References
Adams KL, Wendel JF (2005) Polyploidy and genome evolution in plants. Curr Opin Plant Biol 8:135–141
Alberdi M, Bravo LA, Gutiérrez A, Gidekel M, Corcuera LJ (2002) Ecophysiology of Antarctic vascular plants. Physiol Plant 115:479–486
Albers F (1980) Vergleichende karyologie der Gräser subtriben Aristaveninae und Airinae [Poaceae Aveneae]. Plant Syst Evol 136:137–167
Andreev IO, Spiridonova EV, Kyryachenko SS, Parnikoza IY, Maidanyuk DN, Volkov RA, Kozeretskab IA, Kunakh VA (2010) Population-genetic analysis of Deschampsia antarctica from two regions of maritime antarctica. Moscow Univ Biol Sci Bull 65:208–210
Arohonka T (1982) Chromosome counts of vascular plants of the island Seili in Nauvo, southwestern Finland. Turun Yliopiston Julkaisuja, Sar A 2. Biol Geogr 3:1–12
Barker PF, Filippelli GM, Florindo F, Martin EE, Scher HD (2007) Onset and role of the Antarctic Circumpolar Current. Deep Sea Res Part 2 Top Stud Oceanogr 54:2388–2398
Barros e Silva AE, Guerra M (2010) The meaning of DAPI bands observed after C-banding and FISH procedures. Biotech Histochem 85:115–125
Belyayev A, Raskina O (2013) Chromosome evolution in marginal populations of Aegilops speltoides: causes and consequences. Ann Bot 111:531–538
Bravo LA, Ulloa N, Zuñiga GE, Casanova A, Corcuera LJ, Alberdi M (2001) Cold resistance in Antarctic angiosperms. Physiol Plant 111:55–65
Brochmann C, Brysting AK, Alsos IG, Borgen L, Grundt HH, Scheen AC, Elven R (2004) Polyploidy in arctic plants. Biol J Linn Soc 82:521–536
Byun MY, Lee J, Cui LH, Kang Y, Oh TK, Park H, Lee H, Kim WT (2015) Constitutive expression of DaCBF7, an Antarctic vascular plant Deschampsia antarctica CBF homolog, resulted in improved cold tolerance in transgenic rice plants. Plant Sci 236:61–74
Cardone S, Sawatani P, Rush P, García AM, Poggio L, Schrauf G (2009) Karyological studies in Deschampsia antarctica Desv. (Poaceae). Polar Biol 32:427–433
Chiapella J (2007) A molecular phylogenetic study of Deschampsia (Poaceae: Aveneae) inferred from nuclear ITS and plastid trnL sequence data: support for the recognition of Avenella and Vahlodea. Taxon 56:55–64
Chiapella J, Probatova N (2003) The Deschampsia cespitosa complex (Poaceae: Aveneae) with special reference to Russia. Bot J Linn Soc 142:213–228
Chiapella J, Zuloaga FO (2010) A Revision of Deschampsia, Avenella, and Vahlodea (Poaceae, Poeae, Airinae) in South America. Ann Mo Bot Gard 97:141–162
Chwedorzewska KJ, Giełwanowska I, Szczuka E, Bochenek A (2008) High anatomical and low genetic diversity in Deschampsia antarctica Desv. from King George Island, the Antarctic. Pol Polar Res 29:377–386
Darriba D, Taboada GL, Doallo R, Posada D (2012) jModelTest 2: more models, new heuristics and parallel computing. Nat Methods 9:772
Davlianidze MT (1985) Chromosome numbers in the representatives of the flora from Georgia. Bot Zhurnal 70:698–700 (Moscow & Leningrad)
Di Rienzo JA, Casanoves F, Balzarini MG, Gonzalez L, Tablada M, Robledo CW (2008) InfoStat, versión 2008. Grupo InfoStat, FCA, Universidad Nacional de Córdoba, Argentina
Doyle JJ, Doyle JL (1990) Isolation of plant DNA from fresh tissue. Focus 12:13–15
Dvořák J, Appels R (1982) Chromosome and nucleotide sequence differentiation in genomes of polyploid Triticum species. Theor Appl Genet 63:349–360
Edgar RC (2004) MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797
Engelskjøn T (1979) Chromosome numbers in vascular plants from Norway, including Svalbard. Opera Bot 52:1–38
García-Suárez R, Alonso-Blanco C, Fernandez-Carvajal M, Fernandez-Prieto J, Roca A, Giraldez R (1997) Diversity and systematics of Deschampsia sensu lato (Poaceae), inferred from karyotypes, protein electrophoresis, total genomic DNA hybridization and chloroplast DNA analysis. Plant Syst Evol 205:99–110
Gidekel M, Destefano-Beltran L, García P, Mujica L, Leal P, Cuba M, Fuentes L, Bravo LA, Corcuera LJ, Alberdi M (2003) Identification and characterization of three novel cold acclimation-responsive genes from the extremophile hair grass Deschampsia antarctica Desv. Extremophiles 7:459–469
Greilhuber J, Speta F (1976) C-banded karyotypes in the Scilla hohenackeri group, S. persica, and Puschkinia (Liliaceae). Plant Syst Evol 126:149–188
Guerra M (1988) Introdução à citogenética geral. Guanabara Koogan, Rio de Janeiro
Hagerup O (1932) Über Polyploidie in Beziehung zu Klima, Ökologie und Phylogenie. Hereditas 16:19–40
Harper JA, Thomas ID, Lovatt JA, Thomas HM (2004) Physical mapping of rDNA sites in possible diploid progenitors of polyploid Festuca species. Plant Syst Evol 245:163–168
Hasterok R, Wolny E, Hosiawa M, Kowalczyk M, Kulak-Ksiazczyk S, Ksiazczyk T, Heneen W, Maluszynska J (2006) Comparative analysis of rDNA distribution in chromosomes of various species of Brassicaceae. Ann Bot 97:205–216
Hedberg O (1986) On the manifestation of vivipary in Deschampsia cespitosa s. lat. Symb Bot Ups 27:183–192
Hilu KW (2004) Phylogenetics and chromosomal evolution in the Poaceae (grasses). Aust J Bot 52:13–22
Holderegger R, Stehlik I, Lewis Smith R, Abbott R (2003) Populations of Antarctic hairgrass (Deschampsia antarctica) show low genetic diversity. Arct Antarc Alp Res 35:214–217
Hunziker JH, Stebbins GL (1987) Chromosomal evolution in the Gramineae. In: Soderstrom TR, Hilu KW, Campbell CS, Barkworth ME (eds) Grass systematics and evolution. Smithsonian Institution Press, Washington, pp 179–188
John UP, Polotnianka RM, Sivakumaran KA, Chew O, Mackin L, Kuiper MJ, Talbot JP, Nugent GD, Mautord J, Spangenberg GC (2009) Ice recrystallization inhibition proteins (IRIPs) and freeze tolerance in the cryophilic Antarctic hair grass Deschampsia antarctica E. Desv. Plant Cell Environ 32:336–348
Kawano S (1963) Cytogeography and evolution of the Deschampsia cespitosa complex. Can J Bot 41:719–742
Komárková V, Poncet S, Poncet J (1985) Two native Antarctic vascular plants, Deschampsia antarctica and Colobanthus quitensis: a new southernmost locality and other localities in the Antarctic Peninsula area. Arct Alp Res 17:401–416
Komárková V, Poncet S, Poncet J (1990) Additional and revisited localities of vascular plants Deschampsia antarctica Desv. and Colobanthus quitensis (Kunth) Bartl. in the Antarctic Peninsula area. Arct Alp Res 22:108–113
Książczyk T, Taciak M, Zwierzykowski Z (2010) Variability of ribosomal DNA sites in Festuca pratensis, Lolium perenne, and their intergeneric hybrids, revealed by FISH and GISH. J Appl Genet 51:449–460
Lawrence WE (1945) Some ecotypic relations of Deschampsia cespitosa. Am J Bot 32:298–314
Levin DA (2002) The role of chromosomal change in plant evolution. Oxford University Press, New York
Lideikytė L, Pašakinskienė I, Lemežienė N, Nekrošas S, Kanapeckas J (2008) FISH assessment of ribosomal DNA sites in the chromosome sets of Lolium, Festuca and Festulolium. Agriculture 95:116–124
Löve A, Löve D (1975) IOPB chromosome number reports. Taxon 24:504–507
Maddison WP, Maddison DR (2015) Mesquite: a modular system for evolutionary analysis. Version 3.03. http://mesquiteproject.org. Accessed 19 June 2015
Maluszynska J, Heslop-Harrison J (1993) Physical mapping of rDNA loci in Brassica species. Genome 36:774–781
Montiel P, Smith A, Keiller D (1999) Photosynthetic responses of selected Antarctic plants to solar radiation in the southern maritime Antarctic. Polar Res 18:229–235
Moore DM (1970) Studies in Colobanthus quitensis (Kunth) Bartl. and Deschampsia antarctica Desv. II. Taxonomy, distribution and relationships. Br Antarct Surv Bull 23:63–80
Mosyakin S, Bezusko L, Mosyakin A (2007) Origins of native vascular plants of Antarctica: comments from a historical phytogeography viewpoint. Cytol Genet 41:308–316
Navrotska DO, Twardovska MO, Andreev IO, Parnikoza IY, Betekhtin AA, Zahrychuk OM, Kunakh VA (2014) New forms of chromosome polymorphism in Deschampsia antarctica Desv. from the Argentine islands of the Maritime Antarctic region. Ukr Antarct J 13:185–191
Otto SP, Whitton J (2000) Polyploid incidence and evolution. Annu Rev Genet 34:401–437
Parnikoza IY, Maidanuk D, Kozeretska I (2007) Are Deschampsia antarctica Desv. and Colobanthus quitensis (Kunth) Bartl. Migratory Relicts? Cytol Genet 41:226–229
Paszko B (2006) A critical review and a new proposal of karyotype asymmetry indices. Plant Syst Evol 258:39–48
Petrovsky VV, Zhukova PG (1981) Chromosome numbers and taxonomy of some plant species of Wrangel Island. Bot Zhurnal 66:380–387 (Moscow & Leningrad)
Premoli AC, Mathiasen P, Acosta MC, Ramos VA (2012) Phylogeographically concordant chloroplast DNA divergence in sympatric Nothofagus s.s. How deep can it be? New Phytol 193:261–275
Raskina O, Belyayev A, Nevo E (2004) Quantum speciation in Aegilops: molecular cytogenetic evidence from rDNA cluster variability in natural populations. PNAS 101:14818–14823
Raskina O, Barber JC, Nevo E, Belyayev A (2008) Repetitive DNA and chromosomal rearrangements: Speciation-related events in plant genomes. Cytogenet Genome Res 120:351–357
Reeves A (2001) MicroMeasure: a new computer program for the collection and analysis of cytogenetic data. Genome 44:439–443
Roa F (2011) Análise da distribuição dos sítios de DNA ribossomal 5S e 45S em cariótipos de espécies vegetais. Tese de Doutorado, Programa de Pós-Graduação em Biologia Vegetal Universidade Federal de Pernambuco, Recife
Roa F, Guerra M (2012) Distribution of 45S rDNA sites in chromosomes of plants: structural and evolutionary implications. BMC Evol Biol 12:225
Rohlf FH (2000) NTSYS. Numerical taxonomy and multivariate analysis system. Version 2.1. New York
Romero Zarco C (1986) A new method for estimating karyotype asymmetry. Taxon 35:526–530
Röser M, Winterfeld G, Grebenstein B, Hemleben V (2001) Molecular diversity and physical mapping of 5S rDNA in wild and cultivated oat grasses (Poaceae: Aveneae). Mol Phylogenet Evol 21:198–217
Rothera S, Davy A (1986) Polyploidy and habitat differentiation in Deschampsia cespitosa. New Phytol 102:449–467
Ruhland CT, Day TA (2000) Effects of ultraviolet-B radiation on leaf elongation, production and phenylpropanoid concentrations of Deschampsia antarctica and Colobanthus quitensis in Antarctica. Physiol Plant 109:244–251
Schwarzacher T, Heslop-Harrison P (2000) Practical in situ hybridization. BIOS Scientific Publishers Ltd, Oxford
Schwarzacher T, Ambros P, Schweizer D (1980) Application of Giemsa banding to orchid karyotype analysis. Plant Syst Evol 134:293–297
Schweizer D, Loidl J (1987) A model for heterochromatin dispersion and the evolution of C-band patterns. In: Stahl A, Luciani JM, Vagner-Capodano AM (eds) Chromosomes today 9. Allen and Unwin, London, pp 61–74
Shaw J, Lickey EB, Schilling EE, Small RL (2007) Comparison of whole chloroplast genome sequences to choose noncoding regions for phylogenetic studies in angiosperms: the tortoise and the hare III. Am J Bot 94:275–288
Shelukhina OY, Badaeva E, Loskutov I, Pukhal’Sky V (2007) A comparative cytogenetic study of the tetraploid oat species with the A and C genomes: Avena insularis, A. magna, and A. murphyi. Russ J Genet 43:613–626
Sokolovskaya AP, Probatova NS (1975) Chromosome numbers of some grasses (Poaceae) of the USSR. Bot Zhurnal 60:667–678 (Moscow & Leningrad)
Soltis PS, Soltis DE (2009) The role of hybridization in plant speciation. Annu Rev Plant Biol 60:561–588
Soreng RJ, Peterson PM, Romaschenko K, Davidse G, Zuloaga FO, Judziewicz EJ, Filgueiras TS, Davis JI, Morrone O (2015) A worldwide phylogenetic classification of the Poaceae (Gramineae). J Syst Evol 53:117–137
Stebbins GL (1950) Variation and evolution in plants. Columbia University Press, New York
Stebbins GL (1971) Chromosomal evolution in higher plants. Edward Arnold, London
Stebbins GL (1975) The role of polyploid complexes in the evolution of North American grasslands. Taxon 24:91–106
Swofford DL (2000) PAUP* Phylogenetic Analysis Using Parsimony (* and Other Methods). Version, 4. Sinauer Associates Inc., Sunderland
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729
Tateoka T (1955) Karyotaxonomy in Poaceae III. Further studies of somatic chromosomes. Cytologia 20:296–306
Urdampilleta JD, Coulleri JP, Ferrucci MS, Forni-Martins ER (2013) Karyotype evolution and phylogenetic analyses in the genus Cardiospermum L. (Paullinieae, Sapindaceae). Plant Biol 15:868–881
van de Wouw M, van Dijk P, Huiskes AH (2008) Regional genetic diversity patterns in Antarctic hairgrass (Deschampsia antarctica Desv.). J Biogeogr 35:365–376
Vieira RC, Mantovani A (1995) Anatomia foliar de Deschampsia antarctica Desv. (Gramineae). Rev Bras Bot 18:207–220
Volkov R, Kozeretska I, Kyryachenko S, Andreev I, Maidanyuk D, Parnikoza IY, Kunakh V (2010) Molecular evolution and variability of ITS1–ITS2 in populations of Deschampsia antarctica from two regions of the maritime Antarctic. Polar Sci 4:469–478
Weiss-Schneeweiss H, Emadzade K, Jang TS, Schneeweiss GM (2013) Evolutionary consequences, constraints and potential of polyploidy in plants. Cytogenet Genome Res 140:137–150
Wendel JF (2000) Genome evolution in polyploids. Plant Mol Biol 42:225–249
White TJ, Bruns T, Lee SJWT, Taylor JW (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds) PCR protocols: a guide to methods and applications. Academic Press, New York, pp 315–322
Winterfeld G (2006) Molekularcytogenetische Untersuchungen an Hafergräsern (Aveneae) und Anderen Poaceae. Stapfia 86:1–170
Winterfeld G, Röser M (2007a) Disposition of ribosomal DNAs in the chromosomes of perennial oats (Poaceae: Aveneae). Bot J Linn Soc 155:193–210
Winterfeld G, Röser M (2007b) Chromosomal localization and evolution of satellite DNAs and heterochromatin in grasses (Poaceae), especially tribe Aveneae. Plant Syst Evol 264:75–100
Zoldos V, Papes D, Cerbah M, Panaud O, Besendorfer V, Siljak-Yakovlev S (1999) Molecular-cytogenetic studies of ribosomal genes and heterochromatin reveal conserved genome organization among 11 Quercus species. Theor Appl Genet 99:969–977
Zurita F, Jimenez R, Burgos M, de la Guardia R (1998) Sequential silver staining and in situ hybridization reveal a direct association between rDNA levels and the expression of homologous nucleolar organizing regions: a hypothesis for NOR structure and function. J Cell Sci 111:1433–1439
Acknowledgments
The authors are grateful to CONICET, ANPCyT-FONCyT and SECyT-UNC for financial support and to Dirección Nacional del Antártico and the personnel of the Carlini Station for logistic support for fieldwork in Antarctica. This work was funded by Project PICTO 2010–0095 (ANPCyT-DNA).
Author information
Authors and Affiliations
Corresponding author
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
González, M.L., Urdampilleta, J.D., Fasanella, M. et al. Distribution of rDNA and polyploidy in Deschampsia antarctica E. Desv. in Antarctic and Patagonic populations. Polar Biol 39, 1663–1677 (2016). https://doi.org/10.1007/s00300-016-1890-5
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00300-016-1890-5