Abstract
Key messages
Narrowing down to a single putative target gene behind a leaf senescence mutant and constructing the regulation network by proteomic method.
Abstract
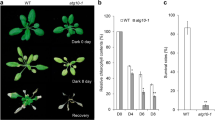
Leaf senescence mutant is an important resource for exploring molecular mechanism of aging. To dig for potential modulation networks during maize leaf aging process, we delimited the gene responsible for a premature leaf senescence mutant els5 to a 1.1 Mb interval in the B73 reference genome using a BC1F1 population with 40,000 plants, and analyzed the leaf proteomics of the mutant and its near-isogenic wild type line. A total of 1355 differentially accumulated proteins (DAP) were mainly enriched in regulation pathways such as “photosynthesis”, “ribosome”, and “porphyrin and chlorophyll metabolism” by the KEGG pathway analysis. The interaction networks constructed by incorporation of transcriptome data showed that ZmELS5 likely repaired several key factors in the photosynthesis system. The putative candidate proteins for els5 were proposed based on DAPs in the fined QTL mapping interval. These results provide fundamental basis for cloning and functional research of the els5 gene, and new insights into the molecular mechanism of leaf senescence in maize.





Similar content being viewed by others
Data availability
The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive in National Genomics Data Center, China National Center for Bioinformation/Beijing Institute of Genomics, Chinese Academy of Sciences, under accession number OMIX002938 that are publicly accessible at https://bigd.big.ac.cn/omix.
References
Asad MAU, Zakari SA, Zhao Q, Zhou L, Ye Y, Cheng F (2019) Abiotic stresses intervene with ABA signaling to induce destructive metabolic pathways leading to death: premature leaf senescence in plants. Int J Mol Sci. https://doi.org/10.3390/ijms20020256
Balazadeh S, Schildhauer J, Araujo WL, Munne-Bosch S, Fernie AR, Proost S, Humbeck K, Mueller-Roeber B (2014) Reversal of senescence by N resupply to N-starved Arabidopsis thaliana: transcriptomic and metabolomic consequences. J Exp Bot 65:3975–3992. https://doi.org/10.1093/jxb/eru119
Chai M, Guo Z, Shi X, Li Y, Tang J, Zhang Z (2019) Dissecting the regulatory network of leaf premature senescence in Maize (Zea mays L.) using transcriptome analysis of ZmELS5 mutant. Genes (Basel). https://doi.org/10.3390/genes10110944
Chen Y, Xu Y, Luo W, Li W, Chen N, Zhang D, Chong K (2013) The F-box protein OsFBK12 targets OsSAMS1 for degradation and affects pleiotropic phenotypes, including leaf senescence, in rice. Plant Physiol 163:1673–1685. https://doi.org/10.1104/pp.113.224527
Chen JH, Wu H, Xu C, Liu XC, Huang Z, Chang S, Wang W, Han G, Kuang T, Shen JR, Zhang X (2020) Architecture of the photosynthetic complex from a green sulfur bacterium. Science. https://doi.org/10.1126/science.abb6350
Chen D, Qiu Z, He L, Hou L, Li M, Zhang G, Wang X, Chen G, Hu J, Gao Z, Dong G, Ren D, Shen L, Zhang Q, Guo L, Qian Q, Zeng D, Zhu L (2021) The rice LRR-like1 protein YELLOW AND PREMATURE DWARF 1 is involved in leaf senescence induced by high light. J Exp Bot 72:1589–1605. https://doi.org/10.1093/jxb/eraa532
Conesa A, Gotz S, Garcia-Gomez JM, Terol J, Talon M, Robles M (2005) Blast2GO: a universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics 21:3674–3676. https://doi.org/10.1093/bioinformatics/bti610
Cui Y, Peng Y, Zhang Q, Xia S, Ruan B, Xu Q, Yu X, Zhou T, Liu H, Zeng D, Zhang G, Gao Z, Hu J, Zhu L, Shen L, Guo L, Qian Q, Ren D (2021) Disruption of EARLY LESION LEAF 1, encoding a cytochrome P450 monooxygenase, induces ROS accumulation and cell death in rice. Plant J 105:942–956. https://doi.org/10.1111/tpj.15079
Diaz C, Purdy S, Christ A, Morot-Gaudry JF, Wingler A, Masclaux-Daubresse C (2005) Characterization of markers to determine the extent and variability of leaf senescence in Arabidopsis. a metabolic profiling approach. Plant Physiol 138:898–908. https://doi.org/10.1104/pp.105.060764
Dods R, Båth P, Morozov D, Gagnér VA, Arlund D, Luk HL, Kübel J, Maj M, Vallejos A, Wickstrand C, Bosman R, Beyerlein KR, Nelson G, Liang M, Milathianaki D, Rbinson J, Harimoorthy R, Berntsen P, Malmerberg E, Johansson L, Andersson R, Carbajo S, Claesson E, Conrad CE, Dahl P, Hammarin G, Hunter MS, Li C, Lisova S, Royant A, Safari C, Sharma A, Williams GJ, Yefanov O, Westenhoff S, Davidsson J, DePonte DP, Boutet S, Barty A, Katona G, Groenhof G, Kübel J, Neutze R (2020) Ultrafast structural changes within a photosynthetic reaction centre. Nature 589:310–314. https://doi.org/10.1038/s41586-020-3000-7
Fincato P, Moschou PN, Ahou A, Angelini R, Roubelakis-Angelakis KA, Federico R, Tavladoraki P (2012) The members of Arabidopsis thaliana PAO gene family exhibit distinct tissue- and organ-specific expression pattern during seedling growth and flower development. Amino Acids 42:831–841. https://doi.org/10.1007/s00726-011-0999-7
Frebort I, Kowalska M, Hluska T, Frebortova J, Galuszka P (2011) Evolution of cytokinin biosynthesis and degradation. J Exp Bot 62:2431–2452. https://doi.org/10.1093/jxb/err004
Guo Y, Gan S (2005) Leaf senescence: signals, execution, and regulation. Curr Top Dev Biol 71:83–112. https://doi.org/10.1016/S0070-2153(05)71003-6
Guo Y, Gan S (2006) AtNAP, a NAC family transcription factor, has an important role in leaf senescence. Plant J 46:601–612. https://doi.org/10.1111/j.1365-313X.2006.02723.x
Guo J, Liu J, Wei Q, Wang R, Yang W, Ma Y, Chen G, Yu Y (2017) Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation in petunias. Plant Physiol 173:668–687. https://doi.org/10.1104/pp.16.00795
Guo Y, Ren G, Zhang K, Li Z, Miao Y, Guo H (2021) Leaf senescence: progression, regulation, and application. Mol Horticulture 1:5. https://doi.org/10.1186/s43897-021-00006-9
Han M, Kim CY, Lee J, Lee SK, Jeon JS (2014) OsWRKY42 represses OsMT1d and induces reactive oxygen species and leaf senescence in rice. Mol Cells 37:532–539. https://doi.org/10.14348/molcells.2014.0128
Hirose N, Takei K, Kuroha T, Kamada-Nobusada T, Hayashi H, Sakakibara H (2008) Regulation of cytokinin biosynthesis, compartmentalization and translocation. J Exp Bot 59:75–83. https://doi.org/10.1093/jxb/erm157
Hong Y, Zhang Y, Sinumporn S, Yu N, Zhan X, Shen X, Chen D, Yu P, Wu W, Liu Q, Cao Z, Zhao C, Cheng S, Cao L (2018) Premature leaf senescence 3, encoding a methyltransferase, is required for melatonin biosynthesis in rice. Plant J. https://doi.org/10.1111/tpj.13995
Hortensteiner S, Feller U (2002) Nitrogen metabolism and remobilization during senescence. J Exp Bot 53:927–937. https://doi.org/10.1093/jexbot/53.370.927
Huang L, Sun Q, Qin F, Li C, Zhao Y, Zhou DX (2007) Down-regulation of a SILENT INFORMATION REGULATOR2-related histone deacetylase gene, OsSRT1, induces DNA fragmentation and cell death in rice. Plant Physiol 144:1508–1519. https://doi.org/10.1104/pp.107.099473
Huang J, Yan M, Zhu X, Zhang T, Shen W, Yu P, Wang Y, Sang X, Yu G, Zhao B, He G (2018) Gene mapping of starch accumulation and premature leaf senescence in the ossac3 mutant of rice. Euphytica 214:177. https://doi.org/10.1007/s10681-018-2261-9
Ile EI, Craufurd PQ, Battey NH, Asiedu R (2006) Phases of dormancy in Yam tubers (Dioscorea rotundata). Ann Bot 97:497–504. https://doi.org/10.1093/aob/mcl002
Jajic I, Sarna T, Strzalka K (2015) Senescence, stress, and reactive oxygen species. Plants (Basel) 4:393–411. https://doi.org/10.3390/plants4030393
Jiang H, Chen Y, Li M, Xu X, Wu G (2011) Overexpression of SGR results in oxidative stress and lesion-mimic cell death in rice seedlings. J Integr Plant Biol 53:375–387. https://doi.org/10.1111/j.1744-7909.2011.01037.x
Kong Z, Li M, Yang W, Xu W, Xue Y (2006) A novel nuclear-localized CCCH-type zinc finger protein, OsDOS, is involved in delaying leaf senescence in rice. Plant Physiol 141:1376–1388. https://doi.org/10.1104/pp.106.082941
Kong W, Yu X, Chen H, Liu L, Xiao Y, Wang Y, Wang C, Lin Y, Yu Y, Wang C, Jiang L, Zhai H, Zhao Z, Wan J (2016) The catalytic subunit of magnesium-protoporphyrin IX monomethyl ester cyclase forms a chloroplast complex to regulate chlorophyll biosynthesis in rice. Plant Mol Biol 92:177–191. https://doi.org/10.1007/s11103-016-0513-4
Koyama T (2014) The roles of ethylene and transcription factors in the regulation of onset of leaf senescence. Front Plant Sci 5:650. https://doi.org/10.3389/fpls.2014.00650
Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M Tanaka A (2007a) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19:1362–1375. https://doi.org/10.1105/tpc.106.042911
Kusaba M, Ito H, Morita R, Iida S, Sato Y, Fujimoto M, Kawasaki S, Tanaka R, Hirochika H, Nishimura M, Tanaka A (2007b) Rice NON-YELLOW COLORING1 is involved in light-harvesting complex II and grana degradation during leaf senescence. Plant Cell 19:1362–1375. https://doi.org/10.1105/tpc.106.042911
Lee S, Seo PJ, Lee HJ, Park CM (2012) A NAC transcription factor NTL4 promotes reactive oxygen species production during drought-induced leaf senescence in Arabidopsis. Plant J 70:831–844. https://doi.org/10.1111/j.1365-313X.2012.04932.x
Lee S, Lee HJ, Huh SU, Paek KH, Ha JH, Park CM (2014) The Arabidopsis NAC transcription factor NTL4 participates in a positive feedback loop that induces programmed cell death under heat stress conditions. Plant Sci 227:76–83. https://doi.org/10.1016/j.plantsci.2014.07.003
Li Z, Zhang Y, Zou D, Zhao Y, Wang HL, Zhang Y, Xia X, Luo J, Guo H, Zhang Z (2020) LSD 3.0: a comprehensive resource for the leaf senescence research community. Nucleic Acids Res 48:D1069–D1075. https://doi.org/10.1093/nar/gkz898
Liang C, Wang Y, Zhu Y, Tang J, Hu B, Liu L, Ou S, Wu H, Sun X, Chu J, Chu C (2014) OsNAP connects abscisic acid and leaf senescence by fine-tuning abscisic acid biosynthesis and directly targeting senescence-associated genes in rice. Proc Natl Acad Sci USA 111:10013–10018. https://doi.org/10.1073/pnas.1321568111
Lim PO, Nam HG (2005) The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr Top Dev Biol 67:49–83. https://doi.org/10.1016/S0070-2153(05)67002-0
Lim PO, Kim HJ, Nam HG (2007) Leaf senescence. Annu Rev Plant Biol 58:115–136. https://doi.org/10.1146/annurev.arplant.57.032905.105316
Lin Y, Tan L, Zhao L, Sun X, Sun C (2016) RLS3, a protein with AAA+ domain localized in chloroplast, sustains leaf longevity in rice. J Integr Plant Biol 58:971–982. https://doi.org/10.1111/jipb.12487
Liu Z, Yan H, Wang K, Kuang T, Zhang J, Gui L, An X, Chang W (2004) Crystal structure of spinach major light-harvesting complex at 2.72 A resolution. Nature 428:287–292. https://doi.org/10.1038/nature02373
Moore B, Zhou L, Rolland F, Hall Q, Cheng WH, Liu YX, Hwang I, Jones T, Sheen J (2003) Role of the Arabidopsis glucose sensor HXK1 in nutrient, light, and hormonal signaling. Science 300:332–336. https://doi.org/10.1126/science.1080585
Morita R, Sato Y, Masuda Y, Nishimura M, Kusaba M (2009) Defect in non-yellow coloring 3, an alpha/beta hydrolase-fold family protein, causes a stay-green phenotype during leaf senescence in rice. Plant J 59:940–952. https://doi.org/10.1111/j.1365-313X.2009.03919.x
Nam HG (1997) The molecular genetic analysis of leaf senescence. Curr Opin Biotechnol 8:200–207. https://doi.org/10.1016/s0958-1669(97)80103-6
Piao W, Han SH, Sakuraba Y, Paek NC (2017) Rice 7-hydroxymethyl chlorophyll a reductase is involved in the promotion of chlorophyll degradation and modulates cell death signaling. Mol Cells 40:773-786. https://doi.org/10.14348/molcells.2017.0127
Qiao Y, Jiang W, Lee J, Park B, Choi MS, Piao R, Woo MO, Roh JH, Han L, Paek NC, Seo HS, Koh HJ (2010) SPL28 encodes a clathrin-associated adaptor protein complex 1, medium subunit micro 1 (AP1M1) and is responsible for spotted leaf and early senescence in rice (Oryza sativa). New Phytol 185:258–274. https://doi.org/10.1111/j.1469-8137.2009.03047.x
Rong H, Tang Y, Zhang H, Wu P, Chen Y, Li M, Wu G, Jiang H (2013) The Stay-Green Rice like (SGRL) gene regulates chlorophyll degradation in rice. J Plant Physiol 170:1367–1373. https://doi.org/10.1016/j.jplph.2013.05.016
Ruan B, Gao Z, Zhao J, Zhang B, Zhang A, Hong K, Yang S, Jiang H, Liu C, Chen G (2017) The rice YGL gene encoding an Mg2+-chelatase ChlD subunit is affected by temperature for chlorophyll biosynthesis. J Plant Biol 60:314–321. https://doi.org/10.1007/s12374-016-0596-0
Sakuraba Y, Balazadeh S, Tanaka R, Mueller-Roeber B, Tanaka A (2012a) Overproduction of chl B retards senescence through transcriptional reprogramming in Arabidopsis. Plant Cell Physiol 53:505–517. https://doi.org/10.1093/pcp/pcs006
Sakuraba Y, Schelbert S, Park SY, Han SH, Lee BD, Andres CB, Kessler F, Hortensteiner S, Paek NC (2012b) STAY-GREEN and chlorophyll catabolic enzymes interact at light-harvesting complex II for chlorophyll detoxification during leaf senescence in Arabidopsis. Plant Cell 24:507–518. https://doi.org/10.1105/tpc.111.089474
Sakuraba Y, Rahman ML, Cho SH, Kim YS, Koh HJ, Yoo SC, Paek NC (2013) The rice faded green leaf locus encodes protochlorophyllide oxidoreductase B and is essential for chlorophyll synthesis under high light conditions. Plant J 74:122–133. https://doi.org/10.1111/tpj.12110
Sato Y, Morita R, Katsuma S, Nishimura M, Tanaka A, Kusaba M (2009) Two short-chain dehydrogenase/reductases, non-yellow coloring 1 and NYC1-LIKE, are required for chlorophyll b and light-harvesting complex II degradation during senescence in rice. Plant J 57:120–131. https://doi.org/10.1111/j.1365-313X.2008.03670.x
Schuller JM, Birrell JA, Tanaka H, Konuma T, Wulfhorst H, Cox N, Schuller SK, Thiemann J, Lubitz W, Setif P, Ikegami T, Engel BD, Kurisu G, Nowaczyk MM (2019) Structural adaptations of photosynthetic complex I enable ferredoxin-dependent electron transfer. Science 363:257–260. https://doi.org/10.1126/science.aau3613
Sheng Z, Lv Y, Li W, Luo R, Wei X, Xie L, Jiao G, Shao G, Wang J, Tang S, Hu P (2017) Yellow-Leaf 1 encodes a magnesium-protoporphyrin IX monomethyl ester cyclase, involved in chlorophyll biosynthesis in rice (Oryza sativa L.). PLoS One 12:e0177989. https://doi.org/10.1371/journal.pone.0177989
Singh S, Giri MK, Singh PK, Siddiqui A, Nandi AK (2013) Down-regulation of OsSAG12-1 results in enhanced senescence and pathogen-induced cell death in transgenic rice plants. J Biosci 38:583–592. https://doi.org/10.1007/s12038-013-9334-7
Song Y, Li C, Zhu Y, Guo P, Wang Q, Zhang L, Wang Z, Di H (2022) Overexpression of ZmIPT2 gene delays leaf senescence and improves grain yield in maize. Front Plant Sci 13:963873. https://doi.org/10.3389/fpls.2022.963873
Su X, Ma J, Wei X, Cao P, Zhu D, Chang W, Liu Z, Zhang X, Li M (2017) Structure and assembly mechanism of plant C2S2M2-type PSII-LHCII supercomplex. Science 357:815-820. https://doi.org/10.1126/science.aan0327
Szklarczyk D, Franceschini A, Wyder S, Forslund K, Heller D, Huerta-Cepas J, Simonovic M, Roth A, Santos A, Tsafou KP, Kuhn M, Bork P, Jensen LJ, von Mering C (2015) STRING v10: protein-protein interaction networks, integrated over the tree of life. Nucl Acids Res 43:D447–D452. https://doi.org/10.1093/nar/gku1003
Tang Y, Li M, Chen Y, Wu P, Wu G, Jiang H (2011) Knockdown of OsPAO and OsRCCR1 cause different plant death phenotypes in rice. J Plant Physiol 168:1952–1959. https://doi.org/10.1016/j.jplph.2011.05.026
Tang K, Lin D, Zheng Q, Liu K, Yang Y, Han Y, Jiao N (2017) Genomic, proteomic and bioinformatic analysis of two temperate phages in Roseobacter clade bacteria isolated from the deep-sea water. BMC Genom 18:485. https://doi.org/10.1186/s12864-017-3886-0
Theis J, Schroda M (2016) Revisiting the photosystem II repair cycle. Plant Signal Behav 11:e1218587. https://doi.org/10.1080/15592324.2016.1218587
Tian X, Ling Y, Fang L, Du P, Sang X, Zhao F, Li Y, Xie R, He G (2013) Gene cloning and functional analysis of yellow green leaf3 (ygl3) gene during the whole-plant growth stage in rice. Genes Genom 35:87–93. https://doi.org/10.1007/s13258-013-0069-5
Uauy C, Distelfeld A, Fahima T, Blechl A, Dubcovsky J (2006) A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314:1298–1301. https://doi.org/10.1126/science.1133649
Wang C, Chen N, Liu J, Jiao P, Liu S, Qu J, Guan S, Ma Y (2022) Overexpression of ZmSAG39 in maize accelerates leaf senescence in Arabidopsis thaliana. Plant Growth Regul 98:451–463. https://doi.org/10.1007/s10725-022-00874-1
Wang M, Zhang T, Peng H, Luo S, Tan J, Jiang K, Heng Y, Zhang X, Guo X, Zheng J, Cheng Z (2018) Rice premature leaf senescence 2, encoding a glycosyltransferase (GT), Is involved in leaf senescence. Front Plant Sci 9:560. https://doi.org/10.3389/fpls.2018.00560
Wang W, Yu LJ, Xu C, Tomizaki T, Zhao S, Umena Y, Chen X, Qin X, Xin Y, Suga M, Han G, Kuang T, Shen JR (2019) Structural basis for blue-green light harvesting and energy dissipation in diatoms. Science. https://doi.org/10.1126/science.aav0365
Wingler A, Purdy S, MacLean JA, Pourtau N (2006) The role of sugars in integrating environmental signals during the regulation of leaf senescence. J Exp Bot 57:391–399. https://doi.org/10.1093/jxb/eri279
Wisniewski JR, Zougman A, Nagaraj N, Mann M (2009) Universal sample preparation method for proteome analysis. Nat Methods 6:359–362. https://doi.org/10.1038/nmeth.1322
Woo HR, Koo HJ, Kim J, Jeong H, Yang JO, Lee IH, Jun JH, Choi SH, Park SJ, Kang B, Kim YW, Phee BK, Kim JH, Seo C, Park C, Kim SC, Park S, Lee B, Lee S, Hwang D, Nam HG, Lim PO (2016) Programming of plant leaf senescence with temporal and inter-organellar coordination of transcriptome in Arabidopsis. Plant Physiol 171:452–467. https://doi.org/10.1104/pp.15.01929
Woo HR, Kim HJ, Lim PO, Nam HG (2019) Leaf senescence: systems and dynamics aspects. Annu Rev Plant Biol 70:347–376. https://doi.org/10.1146/annurev-arplant-050718-095859
Wu L, Ren D, Hu S, Li G, Dong G, Jiang L, Hu X, Ye W, Cui Y, Zhu L, Hu J, Zhang G, Gao Z, Zeng D, Qian Q, Guo L (2016a) Down-regulation of a nicotinate phosphoribosyltransferase gene, OsNaPRT1, leads to withered leaf tips. Plant Physiol 171:1085-1098. https://doi.org/10.1104/pp.15.01898
Wu Z, Zhang X, He B, Diao L, Sheng S, Wang J, Guo X, Su N, Wang L, Jiang L, Wang C, Zhai H, Wan J (2007) A chlorophyll-deficient rice mutant with impaired chlorophyllide esterification in chlorophyll biosynthesis. Plant Physiol 145:29–40. https://doi.org/10.1104/pp.107.100321
Wu X, Ding D, Shi C, Xue Y, Zhang Z, Tang G, Tang J (2016b) microRNA-dependent gene regulatory networks in maize leaf senescence. BMC Plant Biol 16:73. https://doi.org/10.1186/s12870-016-0755-y
Wei X, Su X, Cao P, Liu X, Chang W, Li M, Zhang X, Liu Z (2016) Structure of spinach photosystem II-LHCII supercomplex at 3.2 Å resolution. Nature 534:69-74. https://doi.org/10.1038/nature18020
Xie Q, Liang Y, Zhang J, Zheng H, Dong G, Qian Q, Zuo J (2016) Involvement of a putative bipartite transit peptide in targeting Rice pheophorbide a oxygenase into chloroplasts for chlorophyll degradation during leaf senescence. J Genet Genom 43:145–154. https://doi.org/10.1016/j.jgg.2015.09.012
Xiong E, Li Z, Zhang C, Zhang J, Liu Y, Peng T, Chen Z, Zhao Q (2021) A study of leaf-senescence genes in rice based on a combination of genomics, proteomics and bioinformatics. Brief Bioinform. https://doi.org/10.1093/bib/bbaa305
Xu Y, Gianfagna T, Huang B (2010) Proteomic changes associated with expression of a gene (ipt) controlling cytokinin synthesis for improving heat tolerance in a perennial grass species. J Exp Bot 61:3273–3289. https://doi.org/10.1093/jxb/erq149
Yamatani H, Sato Y, Masuda Y, Kato Y, Morita R, Fukunaga K, Nagamura Y, Nishimura M, Sakamoto W, Tanaka A, Kusaba M (2013) NYC4, the rice ortholog of Arabidopsis THF1, is involved in the degradation of chlorophyll—protein complexes during leaf senescence. Plant J 74:652–662. https://doi.org/10.1111/tpj.12154
Yang Y, Xu J, Huang L, Leng Y, Dai L, Rao Y, Chen L, Wang Y, Tu Z, Hu J, Ren D, Zhang G, Zhu L, Guo L, Qian Q, Zeng D (2016a) PGL, encoding chlorophyllide a oxygenase 1, impacts leaf senescence and indirectly affects grain yield and quality in rice. J Exp Bot 67:1297–1310. https://doi.org/10.1093/jxb/erv529
Yang YL, Xu J, Rao YC, Zeng YJ, Liu HJ, Zheng TT, Zhang GH, Hu J, Guo LB, Qian Q, Zeng DL, Shi QH (2016b) Cloning and functional analysis of pale-green leaf (PGL10) in rice (Oryza sativa L.). Plant Growth Regul 78:69–77. https://doi.org/10.1007/s10725-015-0075-5
Yu N, Liu Q, Zhang Y, Zeng B, Chen Y, Cao Y, Zhang Y, Rani MH, Cheng S, Cao L (2019a) CS3, a Ycf54 domain-containing protein, affects chlorophyll biosynthesis in rice (Oryza sativa L.). Plant Sci 283:11–22. https://doi.org/10.1016/j.plantsci.2019.01.022
Yu T, Lu X, Bai Y, Mei X, Guo Z, Liu C, Cai Y (2019b) Overexpression of the maize transcription factor ZmVQ52 accelerates leaf senescence in Arabidopsis. PLoS One 14:e0221949. https://doi.org/10.1371/journal.pone.0221949
Zhang WY, Xu YC, Li WL, Yang L, Yue X, Zhang XS, Zhao XY (2014) Transcriptional analyses of natural leaf senescence in maize. PLoS One 9:e115617. https://doi.org/10.1371/journal.pone.0115617
Funding
This study was funded by the China Postdoctoral Science Foundation (2020M682295), the National Key Research and Development Program of China (2021YFF100302), and the National Natural Science Foundation of China (32272165).
Author information
Authors and Affiliations
Contributions
Conceptualization, YX, ZF, and JT; data curation, YL, XX, ML, WL; formal analysis, YG, XS, and YC; funding acquisition, YX, XZ, and ZF; investigation, YG, YC, YL, and HL; methodology, YG, XS, and XX; project administration, YX and ZF; resources, WL, XZ, and JT; writing—original draft, YG and YX; writing—review and editing, YX and ZF. All authors have read and agreed to the published version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
The authors declare that there are no conflicts of interest.
Additional information
Communicated by Ajit Kumar Shasany.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gao, Y., Shi, X., Chang, Y. et al. Mapping the gene of a maize leaf senescence mutant and understanding the senescence pathways by expression analysis. Plant Cell Rep 42, 1651–1663 (2023). https://doi.org/10.1007/s00299-023-03051-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-023-03051-4