Abstract
Key message
This article attempts to provide comprehensive review of plant deubiquitinases, paying special attention to recent advances in their biochemical activities and biological functions.
Abstract
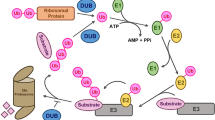
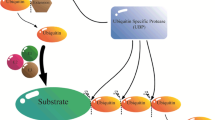
Proteins in eukaryotes are subjected to post-translational modifications, in which ubiquitination is regarded as a reversible process. Cellular deubiquitinases (DUBs) are a key component of the ubiquitin (Ub)–proteasome system responsible for cellular protein homeostasis. DUBs recycle Ub by hydrolyzing poly-Ub chains on target proteins, and maintain a balance of the cellular Ub pool. In addition, some DUBs prefer to cleave poly-Ub chains not linked through the conventional K48 residue, which often alter the substrate activity instead of its stability. In plants, all seven known DUB subfamilies have been identified, namely Ub-binding protease/Ub-specific protease (UBP/USP), Ub C-terminal hydrolase (UCH), Machado–Joseph domain-containing protease (MJD), ovarian-tumor domain-containing protease (OTU), zinc finger with UFM1-specific peptidase domain protease (ZUFSP), motif interacting with Ub-containing novel DUB family (MINDY), and JAB1/MPN/MOV34 protease (JAMM). This review focuses on recent advances in the structure, activity, and biological functions of plant DUBs, particularly in the model plant Arabidopsis.





Similar content being viewed by others
Data availability
The datasets generated and analyzed during the current study are available from the corresponding author on reasonable request.
References
Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, Hofmann K, Kulathu Y (2016) MINDY-1 is a member of an evolutionarily conserved and structurally distinct new family of deubiquitinating enzymes. Mol Cell 63:146–155
Abdul Rehman SA, Armstrong LA, Lange SM, Kristariyanto YA, Grawert TW, Knebel A, Svergun DI, Kulathu Y (2021) Mechanism of activation and regulation of deubiquitinase activity in MINDY1 and MINDY2. Mol Cell 81(4176–4190):e4176
Akutsu M, Dikic I, Bremm A (2016) Ubiquitin chain diversity at a glance. J Cell Sci 129:875–880
Ambroggio XI, Rees DC, Deshaies RJ (2004) JAMM: a metalloprotease-like zinc site in the proteasome and signalosome. PLoS Biol 2:E2
Avila-Ospina L, Moison M, Yoshimoto K, Masclaux-Daubresse C (2014) Autophagy, plant senescence, and nutrient recycling. J Exp Bot 65:3799–3811
Bard JAM, Goodall EA, Greene ER, Jonsson E, Dong KC, Martin A (2018) Structure and function of the 26S proteasome. Annu Rev Biochem 87:697–724
Basters A, Ketscher L, Deuerling E, Arkona C, Rademann J, Knobeloch KP, Fritz G (2012) High yield expression of catalytically active USP18 (UBP43) using a Trigger Factor fusion system. BMC Biotechnol 12:56
Basters A, Geurink PP, El Oualid F, Ketscher L, Casutt MS, Krause E, Ovaa H, Knobeloch KP, Fritz G (2014) Molecular characterization of ubiquitin-specific protease 18 reveals substrate specificity for interferon-stimulated gene 15. FEBS J 281:1918–1928
Basters A, Geurink PP, Rocker A, Witting KF, Tadayon R, Hess S, Semrau MS, Storici P, Ovaa H, Knobeloch KP, Fritz G (2017) Structural basis of the specificity of USP18 toward ISG15. Nat Struct Mol Biol 24:270–278
Basters A, Knobeloch KP, Fritz G (2018) USP18 - a multifunctional component in the interferon response. Biosci Rep 38:BSR20180250. https://doi.org/10.1042/BSR20180250
Bayer RG, Stael S, Csaszar E, Teige M (2011) Mining the soluble chloroplast proteome by affinity chromatography. Proteomics 11:1287–1299
Berndsen CE, Wolberger C (2014) New insights into ubiquitin E3 ligase mechanism. Nat Struct Mol Biol 21:301–307
Bird A (2002) DNA methylation patterns and epigenetic memory. Genes Dev 16:6–21
Block-Schmidt AS, Dukowic-Schulze S, Wanieck K, Reidt W, Puchta H (2011) BRCC36A is epistatic to BRCA1 in DNA crosslink repair and homologous recombination in Arabidopsis thaliana. Nucleic Acids Res 39:146–154
Book AJ, Gladman NP, Lee S-S, Scalf M, Smith LM, Vierstra RD (2010) Affinity purification of the Arabidopsis 26 S proteasome reveals a diverse array of plant proteolytic complexes. J Biol Chem 285:25554–25569
Burnett BG, Pittman RN (2005) The polyglutamine neurodegenerative protein ataxin 3 regulates aggresome formation. P Natl Acad Sci USA 102:4330–4335
Burnett B, Li F, Pittman RN (2003) The polyglutamine neurodegenerative protein ataxin-3 binds polyubiquitylated proteins and has ubiquitin protease activity. Hum Mol Genet 12:3195–3205
Callis J, Raasch JA, Vierstra RD (1990) Ubiquitin extension proteins of Arabidopsis thaliana. Structure, localization, and expression of their promoters in transgenic tobacco. J Biol Chem 265:12486–12493
Callis J, Carpenter T, Sun CW, Vierstra RD (1995) Structure and evolution of genes encoding polyubiquitin and ubiquitin-like proteins in Arabidopsis thaliana ecotype Columbia. Genetics 139:921–939
Cambra I, Martinez M, Dáder B, González-Melendi P, Gandullo J, Santamaría ME, Diaz I (2012) A cathepsin F-like peptidase involved in barley grain protein mobilization, HvPap-1, is modulated by its own propeptide and by cystatins. J Exp Bot 63:4615–4629
Cao S, Engilberge S, Girard E, Gabel F, Franzetti B, Maupin-Furlow JA (2017) Structural insight into ubiquitin-like protein recognition and oligomeric states of JAMM/MPN proteases. Structure 25:823–833
Carpita N, Tierney M, Campbell M (2001) Molecular biology of the plant cell wall: searching for the genes that define structure, architecture and dynamics. Plant Mol Biol 47:1–5
Cartieaux F, Contesto C, Gallou A, Desbrosses G, Kopka J, Taconnat L, Renou JP, Touraine B (2008) Simultaneous interaction of Arabidopsis thaliana with Bradyrhizobium Sp. strain ORS278 and Pseudomonas syringae pv. tomato DC3000 leads to complex transcriptome changes. Mol Plant Microbe Interact 21:244–259
Casal JJ (2012) Shade avoidance. Arabidopsis Book 10:e0157
Catic A, Fiebiger E, Korbel GA, Blom D, Galardy PJ, Ploegh HL (2007) Screen for ISG15-crossreactive deubiquitinases. PLoS One 2:e679
Chamovitz DA (2009) Revisiting the COP9 signalosome as a transcriptional regulator. Embo Rep 10:352–358
Chamovitz DA, Wei N, Osterlund MT, von Arnim AG, Staub JM, Matsui M, Deng XW (1996) The COP9 complex, a novel multisubunit nuclear regulator involved in light control of a plant developmental switch. Cell 86:115–121
Chandler JS, McArdle B, Callis J (1997) AtUBP3 and AtUBP4 are two closely related Arabidopsis thaliana ubiquitin-specific proteases present in the nucleus. Mol Gen Genet 255:302–310
Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R (2020) TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202
Chosed R, Mukherjee S, Lois LM, Orth K (2006) Evolution of a signalling system that incorporates both redundancy and diversity: Arabidopsis SUMOylation. Biochem J 398:521–529
Clague MJ, Barsukov I, Coulson JM, Liu H, Rigden DJ, Urbe S (2013) Deubiquitylases from genes to organism. Physiol Rev 93:1289–1315
Clague MJ, Urbe S, Komander D (2019) Breaking the chains: deubiquitylating enzyme specificity begets function. Nat Rev Mol Cell Biol 20:338–352
Colby T, Matthai A, Boeckelmann A, Stuible HP (2006) SUMO-conjugating and SUMO-deconjugating enzymes from Arabidopsis. Plant Physiol 142:318–332
Cope GA, Suh GSB, Aravind L, Schwarz SE, Zipursky SL, Koonin EV, Deshaies RJ (2002) Role of predicted metalloprotease motif of Jab1/Csn5 in cleavage of Nedd8 from Cul1. Science 298:608–611
Crawford NM (1995) Nitrate: nutrient and signal for plant growth. Plant Cell 7:859–868
Cui X, Lu F, Li Y, Xue Y, Kang Y, Zhang S, Qiu Q, Cui X, Zheng S, Liu B, Xu X, Cao X (2013) Ubiquitin-specific proteases UBP12 and UBP13 act in circadian clock and photoperiodic flowering regulation in Arabidopsis. Plant Physiol 162:897–906
de Jong RN, Ab E, Diercks T, Truffault V, Daniels M, Kaptein R, Folkers GE (2006) Solution structure of the human ubiquitin-specific protease 15 DUSP domain. J Biol Chem 281:5026–5031
Derkacheva M, Liu S, Figueiredo DD, Gentry M, Mozgova I, Nanni P, Tang M, Mannervik M, Kohler C, Hennig L (2016) H2A deubiquitinases UBP12/13 are part of the Arabidopsis polycomb group protein system. Nat Plants 2:16126
Díaz-Mendoza M, Velasco-Arroyo B, González-Melendi P, Martínez M, Díaz I (2014) C1A cysteine protease-cystatin interactions in leaf senescence. J Exp Bot 65:3825–3833
Doelling JH, Yan N, Kurepa J, Walker J, Vierstra RD (2001) The ubiquitin-specific protease UBP14 is essential for early embryo development in Arabidopsis thaliana. Plant J 27:393–405
Doelling JH, Phillips AR, Soyler-Ogretim G, Wise J, Chandler J, Callis J, Otegui MS, Vierstra RD (2007) The ubiquitin-specific protease subfamily UBP3/UBP4 is essential for pollen development and transmission in Arabidopsis. Plant Physiol 145:801–813
Dohmann EMN, Kuhnle C, Schwechheimer C (2005) Loss of the CONSTITUTIVE PHOTOMORPHOGENIC9 signalosome subunit 5 is sufficient to cause the cop/det/fus mutant phenotype in Arabidopsis. Plant Cell 17:1967–1978
Dohmen RJ (2004) SUMO protein modification. Biochim Biophys Acta 1695:113–131
Donaldson KM, Li W, Ching KA, Batalov S, Tsai CC, Joazeiro CAP (2003) Ubiquitin-mediated sequestration of normal cellular proteins into polyglutamine aggregates. P Natl Acad Sci USA 100:8892–8897
Donnison IS, Gay AP, Thomas H, Edwards KJ, Edwards D, James CL, Thomas AM, Ougham HJ (2007) Modification of nitrogen remobilization, grain fill and leaf senescence in maize (Zea mays) by transposon insertional mutagenesis in a protease gene. New Phytol 173:481–494
Drag M, Mikolajczyk J, Bekes M, Reyes-Turcu FE, Ellman JA, Wilkinson KD, Salvesen GS (2008) Positional-scanning fluorigenic substrate libraries reveal unexpected specificity determinants of DUBs (deubiquitinating enzymes). Biochem J 415:367–375
Du L, Li N, Chen L, Xu Y, Li Y, Zhang Y, Li C, Li Y (2014) The ubiquitin receptor DA1 regulates seed and organ size by modulating the stability of the ubiquitin-specific protease UBP15/SOD2 in Arabidopsis. Plant Cell 26:665–677
Du J, Fu L, Sui Y, Zhang L (2020) The function and regulation of OTU deubiquitinases. Front Med 14:542–563
Dzimianski JV, Scholte FEM, Bergeron E, Pegan SD (2019) ISG15: it’s complicated. J Mol Biol 431:4203–4216
Emberley ED, Mosadeghi R, Deshaies RJ (2012) Deconjugation of Nedd8 from Cul1 Is directly regulated by Skp1-F-box and substrate, and the COP9 signalosome inhibits deneddylated SCF by a noncatalytic mechanism. J Biol Chem 287:29679–29689
Ewan R, Pangestuti R, Thornber S, Craig A, Carr C, O’Donnell L, Zhang C, Sadanandom A (2011) Deubiquitinating enzymes AtUBP12 and AtUBP13 and their tobacco homologue NtUBP12 are negative regulators of plant immunity. New Phytol 191:92–106
Franklin KA, Whitelam GC (2005) Phytochromes and shade-avoidance responses in plants. Ann Bot 96:169–175
Frias-Staheli N, Giannakopoulos NV, Kikkert M, Taylor SL, Bridgen A, Paragas J, Richt JA, Rowland RR, Schmaljohn CS, Lenschow DJ, Snijder EJ, García-Sastre A, Virgin HW (2007) Ovarian tumor domain-containing viral proteases evade ubiquitin- and ISG15-dependent innate immune responses. Cell Host Microbe 2:404–416
Gong YY (2016) Identification and characterization of OTU-containing DUBs in Arabidopsis . Capital Normal University
Grasser KD, Rubio V, Barneche F (2021) Multifaceted activities of the plant SAGA complex. Biochim Biophys Acta Gene Regul Mech 1864:194613
Grasty KC, Weeks SD, Loll PJ (2019) Structural insights into the activity and regulation of human Josephin-2. J Struct Biol X 3:100011
Gross CT, McGinnis W (1996) DEAF-1, a novel protein that binds an essential region in a Deformed response element. EMBO J 15:1961–1970
Haahr P, Borgermann N, Guo X, Typas D, Achuthankutty D, Hoffmann S, Shearer R, Sixma TK, Mailand N (2018) ZUFSP deubiquitylates K63-linked polyubiquitin chains to promote genome stability. Mol Cell 70(165–174):e166
Hase Y, Trung KH, Matsunaga T, Tanaka A (2006) A mutation in the uvi4 gene promotes progression of endo-reduplication and confers increased tolerance towards ultraviolet B light. Plant J 46:317–326
Havé M, Marmagne A, Chardon F, Masclaux-Daubresse C (2017) Nitrogen remobilization during leaf senescence: lessons from Arabidopsis to crops. J Exp Bot 68:2513–2529
Hayama R, Yang P, Valverde F, Mizoguchi T, Furutani-Hayama I, Vierstra RD, Coupland G (2019) Ubiquitin carboxyl-terminal hydrolases are required for period maintenance of the circadian clock at high temperature in Arabidopsis. Sci Rep 9:17030
Hermanns T, Pichlo C, Woiwode I, Klopffleisch K, Witting KF, Ovaa H, Baumann U, Hofmann K (2018) A family of unconventional deubiquitinases with modular chain specificity determinants. Nat Commun 9:799
Hewings DS, Heideker J, Ma TP, AhYoung AP, El Oualid F, Amore A, Costakes GT, Kirchhofer D, Brasher B, Pillow T, Popovych N, Maurer T, Schwerdtfeger C, Forrest WF, Yu K, Flygare J, Bogyo M, Wertz IE (2018) Reactive-site-centric chemoproteomics identifies a distinct class of deubiquitinase enzymes. Nat Commun 9:1162
Hofmann K, Falquet L (2001) A ubiquitin-interacting motif conserved in components of the proteasomal and lysosomal protein degradation systems. Trends Biochem Sci 26:347–350
Hofmann RM, Pickart CM (1999) Noncanonical MMS2-encoded ubiquitin-conjugating enzyme functions in assembly of novel polyubiquitin chains for DNA repair. Cell 96:645–653
Hölzl H, Kapelari B, Kellermann J, Seemüller E, Sümegi M, Udvardy A, Medalia O, Sperling J, Müller SA, Engel A, Baumeister W (2000) The regulatory complex of Drosophila melanogaster 26S proteasomes. Subunit composition and localization of a deubiquitylating enzyme. J Cell Biol 150:119–130
Horiguchi G, Kim GT, Tsukaya H (2005) The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43:68–78
Hsieh JWA, Yen MR, Chen PY (2020) Epigenomic regulation of OTU5 in Arabidopsis thaliana. Genomics 112:3549–3559
Isono E, Nagel M-K (2014) Deubiquitylating enzymes and their emerging role in plant biology. Front Plant Sci 5:56
Isono E, Katsiarimpa A, Müller IK, Anzenberger F, Stierhof Y, Geldner N, Chory J, Schwechheimer C (2010) The deubiquitinating enzyme AMSH3 is required for intracellular trafficking and vacuole biogenesis in Arabidopsis thaliana. Plant Cell 22:1826–1837
Jaenisch R, Bird A (2003) Epigenetic regulation of gene expression: how the genome integrates intrinsic and environmental signals. Nat Genet 33:245–254
Jakoby MJ, Falkenhan D, Mader MT, Brininstool G, Wischnitzki E, Platz N, Hudson A, Hülskamp M, Larkin J, Schnittger A (2008) Transcriptional profiling of mature Arabidopsis trichomes reveals that NOECK encodes the MIXTA-like transcriptional regulator MYB106. Plant Physiol 148:1583–1602
Jeong JS, Jung C, Seo JS, Kim JK, Chua NH (2017) The deubiquitinating enzymes UBP12 and UBP13 positively regulate MYC2 levels in jasmonate responses. Plant Cell 29:1406–1424
Jin D, Li B, Deng XW, Wei N (2014) Plant COP9 signalosome subunit 5, CSN5. Plant Sci 224:54–61
Jin D, Wu M, Li B, Bucker B, Keil P, Zhang S, Li J, Kang D, Liu J, Dong J, Deng XW, Irish V, Wei N (2018) The COP9 Signalosome regulates seed germination by facilitating protein degradation of RGL2 and ABI5. PLoS Genet 14:e1007237
Katsiarimpa A, Anzenberger F, Schlager N, Neubert S, Hauser MT, Schwechheimer C, Isono E (2011) The Arabidopsis deubiquitinating enzyme AMSH3 interacts with ESCRT-III subunits and regulates their localization. Plant Cell 23:3026–3040
Katsiarimpa A, Kalinowska K, Anzenberger F, Weis C, Ostertag M, Tsutsumi C, Schwechheimer C, Brunner F, Hückelhoven R, Isono E (2013) The deubiquitinating enzyme AMSH1 and the ESCRT-III subunit VPS2.1 are required for autophagic degradation in Arabidopsis. Plant Cell 25:2236–2252
Katsiarimpa A, Muñoz A, Kalinowska K, Uemura T, Rojo E, Isono E (2014) The ESCRT-III-interacting deubiquitinating enzyme AMSH3 is essential for degradation of ubiquitinated membrane proteins in Arabidopsis thaliana. Plant Cell Physiol 55:727–736
Keller MM, Jaillais Y, Pedmale UV, Moreno JE, Chory J, Ballare CL (2011) Cryptochrome 1 and phytochrome B control shade-avoidance responses in Arabidopsis via partially independent hormonal cascades. Plant J 67:195–207
Keren I, Citovsky V (2017) Activation of gene expression by histone deubiquitinase OTLD1. Epigenetics 12:584–590
Keren I, Lapidot M, Citovsky V (2019) Coordinate activation of a target gene by KDM1C histone demethylase and OTLD1 histone deubiquitinase in Arabidopsis. Epigenetics 14:602–610
Keren I, Citovsky V (2016) The histone deubiquitinase OTLD1 targets euchromatin to regulate plant growth. Sci Signal 9:ra125. https://doi.org/10.1126/scisignal.aaf6767
Keren I, Lacroix B, Kohrman A, Citovsky V (2020) Histone deubiquitinase OTU1 epigenetically regulates DA1 and DA2 which control Arabidopsis seed and organ size. iScience 23:100948
Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25:4877–4887
Komander D, Rape M (2012) The ubiquitin code. Annu Rev Biochem 81:203–229
Komander D, Clague MJ, Urbé S (2009) Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol 10:550–563
Kragelund BB, Schenstrom SM, Rebula CA, Panse VG, Hartmann-Petersen R (2016) DSS1/Sem1, a multifunctional and intrinsically disordered protein. Trends Biochem Sci 41:446–459
Kralemann LEM, Liu SJ, Trejo-Arellano MS, Muñoz-Viana R, Köhler C, Hennig L (2020) Removal of H2Aub1 by ubiquitin-specific proteases 12 and 13 is required for stable Polycomb-mediated gene repression in Arabidopsis. Genome Biol 21:144
Krichevsky A, Zaltsman A, Lacroix B, Citovsky V (2011) Involvement of KDM1C histone demethylase-OTLD1 otubain-like histone deubiquitinase complexes in plant gene repression. Proc Natl Acad Sci USA 108:11157–11162
Kwasna D, Abdul Rehman SA, Natarajan J, Matthews S, Madden R, De Cesare V, Weidlich S, Virdee S, Ahel I, Gibbs-Seymour I, Kulathu Y (2018) Discovery and characterization of ZUFSP/ZUP1, a distinct deubiquitinase class important for genome stability. Mol Cell 70(150–164):e156
Kwok SF, Solano R, Tsuge T, Chamovitz DA, Ecker JR, Matsui M, Deng XW (1998) Arabidopsis homologs of a c-Jun coactivator are present both in monomeric form and in the COP9 complex, and their abundance is differentially affected by the pleiotropic cop/det/fus mutations. Plant Cell 10:1779–1790
Labra M, Ghiani A, Citterio S, Sgorbati S, Sala F, Vannini C, Ruffini-Castiglione M, Bracale M (2002) Analysis of cytosine methylation pattern in response to water deficit in pea root tips. Plant Biol 4:694–699
Lam YA, Xu W, DeMartino GN, Cohen RE (1997) Editing of ubiquitin conjugates by an isopeptidase in the 26S proteasome. Nature 385:737–740
Larsen CN, Krantz BA, Wilkinson KD (1998) Substrate specificity of deubiquitinating enzymes: ubiquitin C-terminal hydrolases. Biochemistry 37:3358–3368
Larson-Rabin Z, Li Z, Masson PH, Day CD (2009) FZR2/CCS52A1 expression is a determinant of endoreduplication and cell expansion in Arabidopsis. Plant Physiol 149:874–884
Lee BL, Singh A, Glover JNM, Hendzel MJ, Spyracopoulos L (2017) Molecular basis for K63-linked ubiquitination processes in double-strand DNA break repair: a focus on kinetics and dynamics. J Mol Biol 429:3409–3429
Li E (2002) Chromatin modification and epigenetic reprogramming in mammalian development. Nat Rev Genet 3:662–673
Li N, Li YH (2016) Signaling pathways of seed size control in plants. Curr Opin Plant Biol 33:23–32
Li T, Duan W, Yang H, Lee MK, Bte Mustafa F, Lee BH, Teo TS (2001) Identification of two proteins, S14 and UIP1, that interact with UCH37. FEBS Lett 488:201–205
Li FS, Macfarlan T, Pittman RN, Chakravarti D (2002) Ataxin-3 is a histone-binding protein with two independent transcriptional corepressor activities. J Biol Chem 277:45004–45012
Li W-F, Perry PJ, Prafulla NN, Schmidt W (2010) Ubiquitin-specific protease 14 (UBP14) is involved in root responses to phosphate deficiency in Arabidopsis. Mol Plant 3:212–223
Li Z, Fu X, Wang Y, Liu R, He Y (2018) Polycomb-mediated gene silencing by the BAH-EMF1 complex in plants. Nat Genet 50:1254–1261
Lindback LN, Hu Y, Ackermann A, Artz O, Pedmale UV (2022) UBP12 and UBP13 deubiquitinases destabilize the CRY2 blue light receptor to regulate Arabidopsis growth. Curr Biol 32(3221–3231):e3226
Liu Y, Wang F, Zhang H, He H, Ma L, Deng XW (2008) Functional characterization of the Arabidopsis ubiquitin-specific protease gene family reveals specific role and redundancy of individual members in development. Plant J 55:844–856
Liu Q, Yan T, Tan X, Wei Z, Li Y, Sun Z, Zhang H, Chen J (2022) Genome-wide identification and gene expression analysis of the OTU DUB family in Oryza sativa. Viruses 14:392
Lu L, Zhai X, Li X, Wang S, Zhang L, Wang L, Jin X, Liang L, Deng Z, Li Z, Wang Y, Fu X, Hu H, Wang J, Mei Z, He Z, Wang F (2022) Met1-specific motifs conserved in OTUB subfamily of green plants enable rice OTUB1 to hydrolyse Met1 ubiquitin chains. Nat Commun 13:4672
Luo M, Luo M-Z, Buzas D, Finnegan J, Helliwell C, Dennis ES, Peacock WJ, Chaudhury A (2008) UBIQUITIN-SPECIFIC PROTEASE 26 is required for seed development and the repression of PHERES1 in Arabidopsis. Genetics 180:229–236
Luo Y, Takagi J, Claus LAN, Zhang C, Yasuda S, Hasegawa Y, Yamaguchi J, Shan L, Russinova E, Sato T (2022) Deubiquitinating enzymes UBP12 and UBP13 stabilize the brassinosteroid receptor BRI1. EMBO Rep 23:e53354
Majumdar P, Nath U (2020) De-ubiquitinases on the move: an emerging field in plant biology. Plant Biol 22:563–572
Majumdar P, Karidas P, Siddiqi I, Nath U (2020) The Ubiquitin-specific protease TNI/UBP14 functions in ubiquitin recycling and affects auxin response. Plant Physiol 184:1499–1513
Makarova KS, Aravind L, Koonin EV (2000) A novel superfamily of predicted cysteine proteases from eukaryotes, viruses and Chlamydia pneumoniae. Trends Biochem Sci 25:50–52
Malakhov MP, Malakhova OA, Kim KI, Ritchie KJ, Zhang DE (2002) UBP43 (USP18) specifically removes ISG15 from conjugated proteins. J Biol Chem 277:9976–9981
Maraschin FD, Memelink J, Offringa R (2009) Auxin-induced, SCFTIR1-mediated poly-ubiquitination marks AUX/IAA proteins for degradation. Plant J 59:100–109
March E, Farrona S (2018) Plant deubiquitinases and their role in the control of gene expression through modification of histones. Front Plant Sci 8:2274
Marechal A, Zou L (2015) RPA-coated single-stranded DNA as a platform for post-translational modifications in the DNA damage response. Cell Res 25:9–23
Maytal-Kivity V, Reis N, Hofmann K, Glickman MH (2002) MPN+, a putative catalytic motif found in a subset of MPN domain proteins from eukaryotes and prokaryotes, is critical for Rpn11 function. BMC Biochem 3:28
Mazzucotelli E, Belloni S, Marone D, De Leonardis A, Guerra D, Di Fonzo N, Cattivelli L, Mastrangelo A (2006) The e3 ubiquitin ligase gene family in plants: regulation by degradation. Curr Genomics 7:509–522
McKenna S, Spyracopoulos L, Moraes T, Pastushok L, Ptak C, Xiao W, Ellison MJ (2001) Noncovalent interaction between ubiquitin and the human DNA repair protein Mms2 is required for Ubc13-mediated polyubiquitination. J Biol Chem 276:40120–40126
Meng LS (2015) Transcription coactivator Arabidopsis ANGUSTIFOLIA3 modulates anthocyanin accumulation and light-induced root elongation through transrepression of Constitutive Photomorphogenic1. Plant Cell Environ 38:838–851
Meng LS, Wang YB, Yao SQ, Liu A (2015a) Arabidopsis AINTEGUMENTA mediates salt tolerance by trans-repressing SCABP8. J Cell Sci 128:2919–2927
Meng LS, Wang ZB, Yao SQ, Liu AZ (2015b) The ARF2-ANT-COR15A gene cascade regulates ABA-signaling-mediated resistance of large seeds to drought in Arabidopsis. J Cell Sci 128:3922–3932
Meng LS, Li YQ, Liu MQ, Jiang JH (2016a) The Arabidopsis ANGUSTIFOLIA3-YODA gene cascade induces anthocyanin accumulation by regulating sucrose levels. Front Plant Sci 7:1728
Meng LS, Wang YB, Loake GJ, Jiang JH (2016b) Seed embryo development is regulated via an gene cascade. Front Plant Sci 7:1645
Meng LS, Xu MK, Li D, Zhou MM, Jiang JH (2017) Soluble sugar accumulation can influence seed size via AN3-YDA gene cascade. J Agr Food Chem 65:4121–4132
Mergner J, Schwechheimer C (2014) The NEDD8 modification pathway in plants. Front Plant Sci 5:103
Metzger MB, Pruneda JN, Klevit RE, Weissman AM (2014) RING-type E3 ligases: master manipulators of E2 ubiquitin-conjugating enzymes and ubiquitination. Biochim Biophys Acta 1843:47–60
Mevissen TET, Komander D (2017) Mechanisms of deubiquitinase specificity and regulation. Annu Rev Biochem 86:159–192
Mevissen TE, Hospenthal MK, Geurink PP, Elliott PR, Akutsu M, Arnaudo N, Ekkebus R, Kulathu Y, Wauer T, El Oualid F, Freund SM, Ovaa H, Komander D (2013) OTU deubiquitinases reveal mechanisms of linkage specificity and enable ubiquitin chain restriction analysis. Cell 154:169–184
Moon BC, Choi MS, Kang YH, Kim MC, Cheong MS, Park CY, Yoo JH, Koo SC, Lee SM, Lim CO, Cho MJ, Chung WS (2005) Arabidopsis ubiquitin-specific protease 6 (AtUBP6) interacts with calmodulin. FEBS Lett 579:3885–3890
Mural RV, Liu Y, Rosebrock TR, Brady JJ, Hamera S, Connor RA, Martin GB, Zeng L (2013) The tomato Fni3 lysine-63-specific ubiquitin-conjugating enzyme and suv ubiquitin E2 variant positively regulate plant immunity. Plant Cell 25:3615–3631
Murtas G, Reeves PH, Fu YF, Bancroft I, Dean C, Coupland G (2003) A nuclear protease required for flowering-time regulation in Arabidopsis reduces the abundance of SMALL UBIQUITIN-RELATED MODIFIER conjugates. Plant Cell 15:2308–2319
Nakada S, Tai I, Panier S, Al-Hakim AK, Iemura SI, Juang YC, O’Donnell L, Kumakubo A, Munro M, Sicheri F, Gingras AC, Natsume T, Suda T, Durocher D (2010) Non-canonical inhibition of DNA damage-dependent ubiquitination by OTUB1. Nature 466:941–946
Narasimhan J, Wang M, Fu Z, Klein JM, Haas AL, Kim JJ (2005) Crystal structure of the interferon-induced ubiquitin-like protein ISG15. J Biol Chem 280:27356–27365
Nassrallah A, Rougee M, Bourbousse C, Drevensek S, Fonseca S, Iniesto E, Ait-Mohamed O, Deton-Cabanillas AF, Zabulon G, Ahmed I, Stroebel D, Masson V, Lombard B, Eeckhout D, Gevaert K, Loew D, Genovesio A, Breyton C, De Jaeger G, Bowler C, Rubio V, Barneche F (2018) DET1-mediated degradation of a SAGA-like deubiquitination module controls H2Bub homeostasis. Elife 7:e37892
Nicastro G, Menon RP, Masino L, Knowles PP, McDonald NQ, Pastore A (2005) The solution structure of the Josephin domain of ataxin-3: structural determinants for molecular recognition. Proc Natl Acad Sci U S A 102:10493–10498
Pamei I, Makandar R (2022) Comparative proteome analysis reveals the role of negative floral regulators and defense-related genes in phytoplasma infected sesame. Protoplasma 259:1441–1453
Pan RH, Kaur N, Hu JP (2014) The Arabidopsis mitochondrial membrane-bound ubiquitin protease UBP27 contributes to mitochondrial morphogenesis. Plant J 78:1047–1059
Park HJ, Kim WY, Park HC, Lee SY, Bohnert HJ, Yun DJ (2011) SUMO and SUMOylation in plants. Mol Cells 32:305–316
Pauwels L, Goossens A (2011) The JAZ proteins: a crucial interface in the jasmonate signaling cascade. Plant Cell 23:3089–3100
Pickart CM (2001) Mechanisms underlying ubiquitination. Annu Rev Biochem 70:503–533
Prins A, van Heerden PD, Olmos E, Kunert KJ, Foyer CH (2008) Cysteine proteinases regulate chloroplast protein content and composition in tobacco leaves: a model for dynamic interactions with ribulose-1,5-bisphosphate carboxylase/oxygenase (Rubisco) vesicular bodies. J Exp Bot 59:1935–1950
Radjacommare R, Usharani R, Kuo CH, Fu H (2014) Distinct phylogenetic relationships and biochemical properties of Arabidopsis ovarian tumor-related deubiquitinases support their functional differentiation. Front Plant Sci 5:84
Rao-Naik C, Chandler JS, McArdle B, Callis J (2000) Ubiquitin-specific proteases from Arabidopsis thaliana: cloning of AtUBP5 and analysis of substrate specificity of AtUBP3, AtUBP4, and AtUBP5 using Escherichia coli in vivo and in vitro assays. Arch Biochem Biophys 379:198–208
Salazar-Henao JE, Vélez-Bermúdez IC, Schmidt W (2016) The regulation and plasticity of root hair patterning and morphogenesis. Development 143:1848–1858
Sato Y, Yoshikawa A, Yamagata A, Mimura H, Yamashita M, Ookata K, Nureki O, Iwai K, Komada M, Fukai S (2008) Structural basis for specific cleavage of Lys 63-linked polyubiquitin chains. Nature 455:358–362
Scaglione KM, Zavodszky E, Todi SV, Patury S, Xu P, Rodriguez-Lebron E, Fischer S, Konen J, Djarmati A, Peng J, Gestwicki JE, Paulson HL (2011) Ube2w and ataxin-3 coordinately regulate the ubiquitin ligase CHIP. Mol Cell 43:599–612
Schaefer JB, Morgan DO (2011) Protein-linked ubiquitin chain structure restricts activity of deubiquitinating enzymes. J Biol Chem 286:45186–45196
Scheel H, Tomiuk S, Hofmann K (2003) Elucidation of ataxin-3 and ataxin-7 function by integrative bioinformatics. Hum Mol Genet 12:2845–2852
Schmitz RJ, Tamada Y, Doyle MR, Zhang XY, Amasino RM (2009) Histone H2B deubiquitination is required for transcriptional activation of FLOWERING LOCUS C and for proper control of flowering in Arabidopsis. Plant Physiol 149:1196–1204
Seeger M, Kraft R, Ferrell K, Bech-Otschir D, Dumdey R, Schade R, Gordon C, Naumann M, Dubiel W (1998) A novel protein complex involved in signal transduction possessing similarities to 26S proteasome subunits. FASEB J 12:469–478
Singer R, Atar S, Atias O, Oron E, Segal D, Hirsch JA, Tuller T, Orian A, Chamovitz DA (2014) Drosophila COP9 signalosome subunit 7 interacts with multiple genomic loci to regulate development. Nucleic Acids Res 42:9761–9770
Singh AK, Chamovitz DA (2019) Role of Cop9 Signalosome subunits in the environmental and hormonal balance of plant. Biomolecules 9:224
Singh AK, Yadav BS, Dhanapal S, Berliner M, Finkelshtein A, Chamovitz DA (2019) CSN5A Subunit of COP9 Signalosome Temporally Buffers Response to Heat in Arabidopsis. Biomolecules 9:805
Singh AK, Dhanapal S, Finkelshtein A, Chamovitz DA (2021) CSN5A subunit of COP9 Signalosome is required for resetting transcriptional stress memory after recurrent heat stress in Arabidopsis. Biomolecules 11(5):668
Song L, Luo ZQ (2019) Post-translational regulation of ubiquitin signaling. J Cell Biol 218:1776–1786
Spoel SH, Mou Z, Tada Y, Spivey NW, Genschik P, Dong X (2009) Proteasome-mediated turnover of the transcription coactivator NPR1 plays dual roles in regulating plant immunity. Cell 137:860–872
Sridhar VV, Kapoor A, Zhang K, Zhu J, Zhou T, Hasegawa PM, Bressan RA, Zhu JK (2007) Control of DNA methylation and heterochromatic silencing by histone H2B deubiquitination. Nature 447:735–738
Stone SL, Arnoldo M, Goring DR (1999) A breakdown of Brassica self-incompatibility in ARC1 antisense transgenic plants. Science 286:1729–1731
Stone M, Hartmann-Petersen R, Seeger M, Bech-Otschir D, Wallace M, Gordon C (2004) Uch2/Uch37 is the major deubiquitinating enzyme associated with the 26S proteasome in fission yeast. J Mol Biol 344:697–706
Suen DF, Tsai YH, Cheng YT, Radjacommare R, Ahirwar RN, Fu H, Schmidt W (2018) The deubiquitinase OTU5 regulates root responses to phosphate starvation. Plant Physiol 176:2441–2455
Sunnerhagen M, Pursglove S, Fladvad M (2002) The new MATH: homology suggests shared binding surfaces in meprin tetramers and TRAF trimers. FEBS Lett 530:1–3
Tian G, Lu Q, Kohalmi SE, Rothstein SJ, Cui Y (2012) Evidence that the Arabidopsis ubiquitin C-terminal hydrolases 1 and 2 associate with the 26S proteasome and the TREX-2 complex. Plant Signal Behav 7:1415–1419
van Bentem SD, Anrather D, Dohnal I, Roitinger E, Csaszar E, Joore J, Buijnink J, Carreri A, Forzani C, Lorkovic ZJ, Barta A, Lecourieux D, Verhounig A, Jonak C, Hirt H (2008) Site-specific phosphorylation profiling of Arabidopsis proteins by mass spectrometry and peptide chip analysis. J Proteome Res 7:2458–2470
Vidal EA, Moyano TC, Krouk G, Katari MS, Tanurdzic M, McCombie WR, Coruzzi GM, Gutierrez RA (2013) Integrated RNA-seq and sRNA-seq analysis identifies novel nitrate-responsive genes in Arabidopsis thaliana roots. BMC Genomics 14:701
Wagner GJ, Wang E, Shepherd RW (2004) New approaches for studying and exploiting an old protuberance, the plant trichome. Ann Bot 93:3–11
Wang Y, Zhang W-Z, Song L-F, Zou J-J, Su Z, Wu W-H (2008) Transcriptome analyses show changes in gene expression to accompany pollen germination and tube growth in Arabidopsis. Plant Physiol 148:1201–1211
Wang F, Zhu DM, Huang X, Li S, Gong YN, Yao QF, Fu XD, Fan LM, Deng XW (2009) Biochemical insights on degradation of Arabidopsis DELLA proteins gained from a cell-free assay system. Plant Cell 21:2378–2390
Wang J, Yu Y, Zhang Z, Quan R, Zhang H, Ma L, Deng XW, Huang R (2013) Arabidopsis CSN5B interacts with VTC1 and modulates ascorbic acid synthesis. Plant Cell 25:625–636
Wang DH, Song W, Wei SW, Zheng YF, Chen ZS, Han JD, Zhang HT, Luo JC, Qin YM, Xu ZH, Bai SN (2018) Characterization of the Ubiquitin C-Terminal hydrolase and Ubiquitin-Specific Protease families in rice (Oryza sativa). Front Plant Sci 9:1636
Wang L, Wen R, Wang J, Xiang D, Wang Q, Zang Y, Wang Z, Huang S, Li X, Datla R, Fobert PR, Wang H, Wei Y, Xiao W (2019) Arabidopsis UBC13 differentially regulates two programmed cell death pathways in responses to pathogen and low-temperature stress. New Phytol 221:919–934
Wang S, Li Q, Zhao L, Fu S, Qin L, Wei Y, Fu YB, Wang H (2020) Arabidopsis UBC22, an E2 able to catalyze lysine-11 specific ubiquitin linkage formation, has multiple functions in plant growth and immunity. Plant Sci 297:110520
Wauer T, Komander D (2014) The JAMM in the proteasome. Nat Struct Mol Biol 21:346–348
Weake VM, Workman JL (2012) SAGA function in tissue-specific gene expression. Trends Cell Biol 22:177–184
Wen R, Newton L, Li G, Wang H, Xiao W (2006) Arabidopsis thaliana UBC13: implication of error-free DNA damage tolerance and Lys63-linked polyubiquitylation in plants. Plant Mol Biol 61:241–253
Wen R, Torres-Acosta JA, Pastushok L, Lai X, Pelzer L, Wang H, Xiao W (2008) Arabidopsis UEV1D promotes lysine-63 linked polyubiquitination and is involved in DNA damage response. Plant Cell 20:213–227
Wiener R, Zhang X, Wang T, Wolberger C (2012) The mechanism of OTUB1-mediated inhibition of ubiquitination. Nature 483:618–622
Winborn BJ, Travis SM, Todi SV, Scaglione KM, Xu P, Williams AJ, Cohen RE, Peng J, Paulson HL (2008) The deubiquitinating enzyme ataxin-3, a polyglutamine disease protein, edits Lys63 linkages in mixed linkage ubiquitin chains. J Biol Chem 283:26436–26443
Worden EJ, Padovani C, Martin A (2014) Structure of the Rpn11-Rpn8 dimer reveals mechanisms of substrate deubiquitination during proteasomal degradation. Nat Struct Mol Biol 21:220–227
Xu D (2020) COP1 and BBXs-HY5-mediated light signal transduction in plants. New Phytol 228:1748–1753
Xu Y, Jin W, Li N, Zhang W, Liu C, Li C, Li Y (2016) UBIQUITIN-SPECIFIC PROTEASE14 interacts with ULTRAVIOLET-B INSENSITIVE4 to regulate endoreduplication and cell and organ growth in Arabidopsis. Plant Cell 28:1200–1214
Yan N, Doelling JH, Falbel TG, Durski AM, Vierstra RD (2000) The ubiquitin-specific protease family from Arabidopsis. AtUBP1 and 2 are required for the resistance to the amino acid analog canavanine. Plant Physiol 124:1828–1843
Yang P, Smalle J, Lee SS, Yan N, Emborg TJ, Vierstra RD (2007) Ubiquitin C-terminal hydrolases 1 and 2 affect shoot architecture in Arabidopsis. Plant J 51:441–457
Yao D, Arguez MA, He P, Bent AF, Song J (2021) Coordinated regulation of plant immunity by poly(ADP-ribosyl)ation and K63-linked ubiquitination. Mol Plant 14:208–2103
Ye H, Park YC, Kreishman M, Kieff E, Wu H (1999) The structural basis for the recognition of diverse receptor sequences by TRAF2. Mol Cell 4:321–330
Ye Y, Akutsu M, Reyes-Turcu F, Enchev RI, Wilkinson KD, Komander D (2011) Polyubiquitin binding and cross-reactivity in the USP domain deubiquitinase USP21. EMBO Rep 12:350–357
Ye Y, Blaser G, Horrocks MH, Ruedas-Rama MJ, Ibrahim S, Zhukov AA, Orte A, Klenerman D, Jackson SE, Komander D (2012) Ubiquitin chain conformation regulates recognition and activity of interacting proteins. Nature 492:266–270
Yen MR, Suen DF, Hsu FM, Tsai YH, Fu H, Schmidt W, Chen PY (2017) Deubiquitinating enzyme OTU5 contributes to DNA methylation patterns and is critical for phosphate nutrition signals. Plant Physiol 175:1826–1838
Yin XJ, Volk S, Ljung K, Mehlmer N, Dolezal K, Ditengou F, Hanano S, Davis SJ, Schmelzer E, Sandberg G, Teige M, Palme K, Pickart C, Bachmair A (2007) Ubiquitin lysine 63 chain forming ligases regulate apical dominance in Arabidopsis. Plant Cell 19:1898–1911
Young P, Deveraux Q, Beal RE, Pickart CM, Rechsteiner M (1998) Characterization of two polyubiquitin binding sites in the 26 S protease subunit 5a. J Biol Chem 273:5461–5467
Yu X, Klejnot J, Zhao X, Shalitin D, Maymon M, Yang H, Lee J, Liu X, Lopez J, Lin C (2007) Arabidopsis cryptochrome 2 completes its posttranslational life cycle in the nucleus. Plant Cell 19:3146–3156
Zang Y, Gong Y, Wang Q, Guo H, Xiao W (2020) Arabidopsis OTU1, a linkage-specific deubiquitinase, is required for endoplasmic reticulum-associated protein degradation. Plant J 101:141–155
Zeng C, Zhao C, Ge F, Li Y, Cao J, Ying M, Lu J, He Q, Yang B, Dai X, Zhu H (2020) Machado-Joseph deubiquitinases: from cellular functions to potential therapy targets. Front Pharmacol 11:1311
Zhang X, Oppenheimer DG (2004) A simple and efficient method for isolating trichomes for downstream analyses. Plant Cell Physiol 45:221–224
Zhao J, Zhou H, Li Y (2013) UBIQUITIN-SPECIFIC PROTEASE16 interacts with a HEAVY METAL ASSOCIATED ISOPRENYLATED PLANT PROTEIN27 and modulates cadmium tolerance. Plant Signal Behav 8:e25680
Zhao J, Zhou H, Zhang M, Gao Y, Li L, Gao Y, Li M, Yang Y, Guo Y, Li X (2016) Ubiquitin-specific protease 24 negatively regulates abscisic acid signalling in Arabidopsis thaliana. Plant Cell Environ 39:427–440
Zheng N, Shabek N (2017) Ubiquitin ligases: structure, function, and regulation. Annu Rev Biochem 86:129–157
Zheng N, Wang P, Jeffrey PD, Pavletich NP (2000) Structure of a c-Cbl-UbcH7 complex: RING domain function in ubiquitin-protein ligases. Cell 102:533–539
Zhou H, Zhao J, Yang Y, Chen C, Liu Y, Jin X, Chen L, Li X, Deng X, W., Schumaker KS, Guo Y, (2012) Ubiquitin-specific protease16 modulates salt tolerance in Arabidopsis by regulating Na(+)/H(+) antiport activity and serine hydroxymethyltransferase stability. Plant Cell 24:5106–5122
Zhou Y, Romero-Campero FJ, Gomez-Zambrano A, Turck F, Calonje M (2017) H2A monoubiquitination in Arabidopsis thaliana is generally independent of LHP1 and PRC2 activity. Genome Biol 18:69
Zhou Y, Park SH, Soh MY, Chua NH (2021) Ubiquitin-specific proteases UBP12 and UBP13 promote shade avoidance response by enhancing PIF7 stability. Proc Natl Acad Sci USA 118:e2103633118
Acknowledgements
The authors wish to thank Yingya Gong and Yuepeng Zang for sharing unpublished observations and other Xiao laboratory members for discussion.
Funding
This work was supported by the Capital Normal University and a Saskatchewan Canola Development Commission grant ADF 20210893 to WX.
Author information
Authors and Affiliations
Contributions
RL and WX conceived and designed research. RL and KY collected and analyzed data. LW, KY and WX wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no known competing financial and personal relationship conflicts.
Additional information
Communicated by Wusheng Liu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Luo, R., Yang, K. & Xiao, W. Plant deubiquitinases: from structure and activity to biological functions. Plant Cell Rep 42, 469–486 (2023). https://doi.org/10.1007/s00299-022-02962-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-022-02962-y