Abstract
Key message
J-like proteins (JLPs) are emerging as ancillaries to the cellular chaperone network. They modulate functions of Hsp70:J-domain protein (JDP) systems in novel ways thereby having key roles in diverse plant processes.
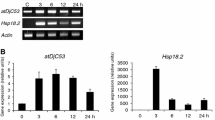
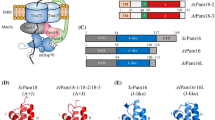
J-domain proteins (JDPs) form an obligate co-chaperone partnership with Hsp70s with their highly conserved J-domain to steer protein quality control processes in the cell. The HPD motif between helix II and helix III of the J-domain is crucial for JDP’s interaction with Hsp70s. According to the most recent classification, J-like proteins (JLPs) form an extended class of the JDP family possessing a degenerate J-domain with the HPD motif non-conservatively replaced by other amino acid residues and hence are not able to interact with Hsp70s. Considering this most updated and acceptable JLP classification, we identified 21 JLPs in Arabidopsis thaliana that share a structurally conserved J-like domain (JLD), but lack the HPD motif. Analysis of publicly available gene expression data as well as real-time quantitative PCR performed for a few selected JLPs implicated some of these proteins in growth, development and stress response. Here, we summarize the current state of knowledge on plant JLPs and their involvement in vital plant cellular/metabolic processes, including chloroplast division, mitochondrial protein import and flowering. Finally, we propose possible modes of action for these highly elusive proteins and other DnaJ-related proteins (DNAJRs) in regulating the Hsp70 chaperone network.




Similar content being viewed by others
References
Ajit Tamadaddi C, Sahi C (2016) J domain independent functions of J proteins. Cell Stress Chaperones 21:563–570
Ajjawi I, Coku A, Froehlich JE, Yang Y, Osteryoung KW, Benning C, Last RL (2011) A J-like protein influences fatty acid composition of chloroplast lipids in Arabidopsis. PLoS ONE 6:e25368
Berardini TZ, Reiser L, Li D, Mezheritsky Y, Muller R, Strait E, Huala E (2015) The Arabidopsis information resource: making and mining the “gold standard” annotated reference plant genome. Genesis (new York, NY, 2000) 53:474–485
Blatch GL, Lässle M (1999) The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays 21:932–939
Cao M, Wei C, Zhao L, Wang J, Jia Q, Wang X, Jin Q, Deng T (2014) DnaJA1/Hsp40 is co-opted by influenza A virus to enhance its viral RNA polymerase activity. J Virol 88:14078–14089
Chen X, Ghazanfar B, Khan AR, Hayat S, Cheng Z (2013) Comparative analysis of putative orthologues of mitochondrial import motor subunit: Pam18 and Pam16 in plants. PLoS ONE 8:e78400
Ciou HS, Tsai YL, Chiu CC (2020) Arabidopsis chloroplast J protein DJC75/CRRJ mediates nitrate-promoted seed germination in the dark. Ann Bot 125:1091–1099
Corpet F (1988) Multiple sequence alignment with hierarchical clustering. Nucleic Acids Res 16:10881–10890
D’Silva PR, Schilke B, Walter W, Craig EA (2005) Role of Pam16’s degenerate J domain in protein import across the mitochondrial inner membrane. Proc Natl Acad Sci USA 102:12419–12424
D’Silva PR, Schilke B, Hayashi M, Craig EA (2008) Interaction of the J-protein heterodimer Pam18/Pam16 of the mitochondrial import motor with the translocon of the inner membrane. Mol Biol Cell 19:424–432
Ducett JK, Peterson FC, Hoover LA, Prunuske AJ, Volkman BF, Craig EA (2013) Unfolding of the C-terminal domain of the J-protein Zuo1 releases autoinhibition and activates Pdr1-dependent transcription. J Mol Biol 425:19–31
Feng P, Lin H, Chen W, Zuo Y (2014) Predicting the types of J-proteins using clustered amino acids. BioMed Res Int 2014:935719
Finka A, Mattoo RU, Goloubinoff P (2011) Meta-analysis of heat- and chemically upregulated chaperone genes in plant and human cells. Cell Stress Chaperones 16:15–31
Frazier AE, Dudek J, Guiard B, Voos W, Li Y, Lind M, Meisinger C, Geissler A, Sickmann A, Meyer HE, Bilanchone V, Cumsky MG, Truscott KN, Pfanner N, Rehling P (2004) Pam16 has an essential role in the mitochondrial protein import motor. Nat Struct Mol Biol 11:226–233
Gillis J, Schipper-Krom S, Juenemann K, Gruber A, Coolen S, van den Nieuwendijk R, van Veen H, Overkleeft H, Goedhart J, Kampinga HH, Reits EA (2013) The DNAJB6 and DNAJB8 protein chaperones prevent intracellular aggregation of polyglutamine peptides. J Biol Chem 288:17225–17237
Glynn JM, Froehlich JE, Osteryoung KW (2008) Arabidopsis ARC6 coordinates the division machineries of the inner and outer chloroplast membranes through interaction with PDV2 in the intermembrane space. Plant Cell 20:2460–2470
Glynn JM, Yang Y, Vitha S, Schmitz AJ, Hemmes M, Miyagishima SY, Osteryoung KW (2009) PARC6, a novel chloroplast division factor, influences FtsZ assembly and is required for recruitment of PDV1 during chloroplast division in Arabidopsis. Plant J 59:700–711
Goodacre NF, Gerloff DL, Uetz P (2013) Protein domains of unknown function are essential in bacteria. Mbio 5:e00744-00713
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
Hageman J, Kampinga HH (2009) Computational analysis of the human HSPH/HSPA/DNAJ family and cloning of a human HSPH/HSPA/DNAJ expression library. Cell Stress Chaperones 14:1–21
Harris CJ, Scheibe M, Wongpalee SP, Liu W, Cornett EM, Vaughan RM, Li X, Chen W, Xue Y, Zhong Z, Yen L, Barshop WD, Rayatpisheh S, Gallego-Bartolome J, Groth M, Wang Z, Wohlschlegel JA, Du J, Rothbart SB, Butter F, Jacobsen SE (2018) A DNA methylation reader complex that enhances gene transcription. Science 362:1182–1186
Hennessy F, Cheetham ME, Dirr HW, Blatch GL (2000) Analysis of the levels of conservation of the J domain among the various types of DnaJ-like proteins. Cell Stress Chaperones 5:347–358
Hennessy F, Nicoll WS, Zimmermann R, Cheetham ME, Blatch GL (2005) Not all J domains are created equal: implications for the specificity of Hsp40-Hsp70 interactions. Protein Sci 14:1697–1709
Huang Y, Chen X, Liu Y, Roth C, Copeland C, McFarlane HE, Huang S, Lipka V, Wiermer M, Li X (2013) Mitochondrial AtPAM16 is required for plant survival and the negative regulation of plant immunity. Nat Commun 4:2558
Jiang Y, Rossi P, Kalodimos CG (2019) Structural basis for client recognition and activity of Hsp40 chaperones. Science 365:1313–1319
Kampinga HH, Craig EA (2010) The HSP70 chaperone machinery: J proteins as drivers of functional specificity. Nat Rev Mol Cell Biol 11:579–592
Kampinga HH, Andreasson C, Barducci A, Cheetham ME, Cyr D, Emanuelsson C, Genevaux P, Gestwicki JE, Goloubinoff P, Huerta-Cepas J, Kirstein J, Liberek K, Mayer MP, Nagata K, Nillegoda NB, Pulido P, Ramos C, De Los RP, Rospert S, Rosenzweig R, Sahi C, Taipale M, Tomiczek B, Ushioda R, Young JC, Zimmermann R, Zylicz A, Zylicz M, Craig EA, Marszalek J (2019) Function, evolution, and structure of J-domain proteins. Cell Stress Chaperones 24:7–15
Kawai-Yamada M, Saito Y, Jin L, Ogawa T, Kim KM, Yu LH, Tone Y, Hirata A, Umeda M, Uchimiya H (2005) A novel Arabidopsis gene causes Bax-like lethality in Saccharomyces cerevisiae. J Biol Chem 280:39468–39473
Knox C, Luke GA, Blatch GL, Pesce E-R (2011) Heat shock protein 40 (Hsp40) plays a key role in the virus life cycle. Virus Res 160:15–24
Kradolfer D, Wolff P, Jiang H, Siretskiy A, Köhler C (2013) An imprinted gene underlies postzygotic reproductive isolation in Arabidopsis thaliana. Dev Cell 26:525–535
Kufareva I, Abagyan R (2012) Methods of protein structure comparison. Methods Mol Biol 857:231–257
Lee JY, Lee HS, Song JY, Jung YJ, Reinbothe S, Park YI, Lee SY, Pai HS (2013) Cell growth defect factor1/chaperone-like protein of POR1 plays a role in stabilization of light-dependent protochlorophyllide oxidoreductase in Nicotiana benthamiana and Arabidopsis. Plant Cell 25:3944–3960
Lee H-S, Choi I, Jeon Y, Ahn H-K, Cho H, Kim J, Kim J-H, Lee J-M, Lee S, Bünting J, Seo DH, Lee T, Lee D-H, Lee I, Oh M-H, Kim T-W, Belkhadir Y, Pai H-S (2021) Chaperone-like protein DAY plays critical roles in photomorphogenesis. Nat Commun 12:4194
Letunic I, Bork P (2018) 20 years of the SMART protein domain annotation resource. Nucleic Acids Res 46:D493-d496
Li Y, Dudek J, Guiard B, Pfanner N, Rehling P, Voos W (2004) The presequence translocase-associated protein import motor of mitochondria. Pam16 functions in an antagonistic manner to Pam18. J Biol Chem 279:38047–38054
Liu Q, Liang C, Zhou L (2020) Structural and functional analysis of the Hsp70/Hsp40 chaperone system. Protein Sci 29:378–390
Lu Z, Cyr DM (1998) The conserved carboxyl terminus and zinc finger-like domain of the co-chaperone Ydj1 assist Hsp70 in protein folding. J Biol Chem 273:5970–5978
Martin JL (1995) Thioredoxin—a fold for all reasons. Structure 3:245–250
Mehawej C, Delahodde A, Legeai-Mallet L, Delague V, Kaci N, Desvignes JP, Kibar Z, Capo-Chichi JM, Chouery E, Munnich A, Cormier-Daire V, Mégarbané A (2014) The impairment of MAGMAS function in human is responsible for a severe skeletal dysplasia. PLoS Genet 10:e1004311
Mokranjac D, Sichting M, Popov-Celeketic D, Berg A, Hell K, Neupert W (2005) The import motor of the yeast mitochondrial TIM23 preprotein translocase contains two different J proteins, Tim14 and Mdj2. J Biol Chem 280:31608–31614
Mokranjac D, Bourenkov G, Hell K, Neupert W, Groll M (2006) Structure and function of Tim14 and Tim16, the J and J-like components of the mitochondrial protein import motor. EMBO J 25:4675–4685
Nillegoda NB, Kirstein J, Szlachcic A, Berynskyy M, Stank A, Stengel F, Arnsburg K, Gao X, Scior A, Aebersold R, Guilbride DL, Wade RC, Morimoto RI, Mayer MP, Bukau B (2015) Crucial HSP70 co-chaperone complex unlocks metazoan protein disaggregation. Nature 524:247–251
Nillegoda NB, Stank A, Malinverni D, Alberts N, Szlachcic A, Barducci A, De Los RP, Wade RC, Bukau B (2017) Evolution of an intricate J-protein network driving protein disaggregation in eukaryotes. Elife 6:e24560
Park HY, Lee SY, Seok HY, Kim SH, Sung ZR, Moon YH (2011) EMF1 interacts with EIP1, EIP6 or EIP9 involved in the regulation of flowering time in Arabidopsis. Plant Cell Physiol 52:1376–1388
Pulido P, Leister D (2018) Novel DNAJ-related proteins in Arabidopsis thaliana. New Phytol 217:480–490
Rajan VB, D’Silva P (2009) Arabidopsis thaliana J-class heat shock proteins: cellular stress sensors. Funct Integr Genomics 9:433–446
Rebeaud ME, Mallik S, Goloubinoff P, Tawfik DS (2021) On the evolution of chaperones and cochaperones and the expansion of proteomes across the Tree of Life. Proc Natl Acad Sci 118:e2020885118
Ratheesh KR, Nagarajan NS, Arunraj SP, Sinha D, Rajan VBV, Esthaki VK, D’Silva P (2012) HSPIR: a manually annotated heat shock protein information resource. Bioinformatics 28:2853–2855
Sagarika P, Dobriyal N, Sahi C (2021) Dosage sensitivity of JDPs, a valuable tool for understanding their function: a case study on Caj1 overexpression-mediated filamentous growth in budding yeast. Curr Genet
Sahi C, Lee T, Inada M, Pleiss JA, Craig EA (2010) Cwc23, an essential J protein critical for pre-mRNA splicing with a dispensable J domain. Mol Cell Biol 30:33–42
Sha B, Lee S, Cyr DM (2000) The crystal structure of the peptide-binding fragment from the yeast Hsp40 protein Sis1. Structure 8:799–807
Shen Y, Hendershot LM (2005) ERdj3, a stress-inducible endoplasmic reticulum DnaJ homologue, serves as a cofactor for BiP’s interactions with unfolded substrates. Mol Biol Cell 16:40–50
Sinha D, Srivastava S, D’Silva P (2016) Functional diversity of human mitochondrial J-proteins is independent of their association with the inner membrane presequence translocase. J Biol Chem 291:17345–17359
Szabo A, Korszun R, Hartl FU, Flanagan J (1996) A zinc finger-like domain of the molecular chaperone DnaJ is involved in binding to denatured protein substrates. EMBO J 15:408–417
Tamadaddi C, Sagar V, Verma AK, Afsal F, Sahi C (2021) Expansion of the evolutionarily conserved network of J-domain proteins in the Arabidopsis mitochondrial import complex. Plant Mol Biol 105:385–403
Verges E, Colomina N, Gari E, Gallego C, Aldea M (2007) Cyclin Cln3 is retained at the ER and released by the J chaperone Ydj1 in late G1 to trigger cell cycle entry. Mol Cell 26:649–662
Verma AK, Tamadaddi C, Tak Y, Lal SS, Cole SJ, Hines JK, Sahi C (2019) The expanding world of plant J-domain proteins. Crit Rev Plant Sci 38:382–400
Walsh P, Bursac D, Law YC, Cyr D, Lithgow T (2004) The J-protein family: modulating protein assembly, disassembly and translocation. EMBO Rep 5:567–571
Waterhouse A, Bertoni M, Bienert S, Studer G, Tauriello G, Gumienny R, Heer FT, de Beer TAP, Rempfer C, Bordoli L, Lepore R, Schwede T (2018) SWISS-MODEL: homology modelling of protein structures and complexes. Nucleic Acids Res 46:W296-w303
Yamamoto H, Peng L, Fukao Y, Shikanai T (2011) An Src homology 3 domain-like fold protein forms a ferredoxin binding site for the chloroplast NADH dehydrogenase-like complex in Arabidopsis. Plant Cell 23:1480–1493
Yang C, Compton MM, Yang P (2005) Dimeric novel HSP40 is incorporated into the radial spoke complex during the assembly process in flagella. Mol Biol Cell 16:637–648
Zhang B, Qiu HL, Qu DH, Ruan Y, Chen DH (2018) Phylogeny-dominant classification of J-proteins in Arabidopsis thaliana and Brassica oleracea. Genome 61:405–415
Zhou K, Ren Y, Lv J, Wang Y, Liu F, Zhou F, Zhao S, Chen S, Peng C, Zhang X, Guo X, Cheng Z, Wang J, Wu F, Jiang L, Wan J (2013) Young leaf chlorosis 1, a chloroplast-localized gene required for chlorophyll and lutein accumulation during early leaf development in rice. Planta 237:279–292
Acknowledgements
We sincerely thank all the laboratory members for their valuable discussions on this manuscript. CT and AV thank the Department of Science and Technology (DST) INSPIRE fellowship for Ph.D. and Kishore Vaigyanik Protsahan Yojana (KVPY) scholarship for undergraduate study and, respectively.
Funding
This work was supported by the Science and Engineering Research Board (SERB-EMR/2015/001213) and Department of Biotechnology, Government of India (BT/PR12149/BRB/10/1348/2014), and intramural funds from IISER Bhopal to CS.
Author information
Authors and Affiliations
Contributions
CT, AKV, and CS conceived the idea; CT and DD performed the qRT-PCR experiments and analysed the data. AKV, CT, VZ, and AV performed the bioinformatics studies. CT, AKV, VZ, and CS wrote the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
We have no conflicts of interest to declare.
Additional information
Communicated by Wusheng Liu.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Tamadaddi, C., Verma, A.K., Zambare, V. et al. J-like protein family of Arabidopsis thaliana: the enigmatic cousins of J-domain proteins. Plant Cell Rep 41, 1343–1355 (2022). https://doi.org/10.1007/s00299-022-02857-y
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-022-02857-y