Abstract
Key message
We identified FLY as a previously uncharacterized RNA-binding-family protein that controls flowering time by positively regulating the expression of FLC clade members.
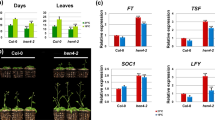
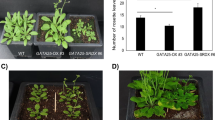
The ability of flowering plants to adjust the timing of the floral transition based on endogenous and environmental signals contributes to their adaptive success. In Arabidopsis thaliana, the MADS-domain protein FLOWERING LOCUS C (FLC) and the FLC clade members FLOWERING LOCUS M/MADS AFFECTING FLOWERING1 (FLM/MAF1), MAF2, MAF3, MAF4, and MAF5 form nuclear complexes that repress flowering under noninductive conditions. However, how FLM/MAF genes are regulated requires further study. Using a genetic strategy, we showed that the previously uncharacterized K-homology (KH) domain protein FLOWERING LOCUS Y (FLY) modulates flowering time. The fly-1 knockout mutant and FLY artificial microRNA knockdown line flowered earlier than the wild type under long- and short-day conditions. The knockout fly-1 allele, a SALK T-DNA insertion mutant, contains an ~ 110-kb genomic deletion induced by T-DNA integration. FLC clade members were downregulated in the fly-1 mutants and FLY artificial microRNA knockdown line, whereas the level of the FLC antisense transcript COOLAIR was similar to that of the wild type. Our results identify FLY as a regulator that affects flowering time through upregulation of FLC clade members.





Similar content being viewed by others
Abbreviations
- FLC:
-
FLOWERING LOCUS C
- FLM:
-
FLOWERING LOCUS M
- FLY:
-
FLOWERING LOCUS Y
- HEN4:
-
HUA ENHANCER 4
- KH:
-
K-homology
- MAF:
-
MADS AFFECTING FLOWERING
References
Airoldi CA, McKay M, Davies B (2015) MAF2 is regulated by temperature-dependent splicing and represses flowering at low temperatures in parallel with FLM. PLoS ONE 10:e0126516
Bouche F, Woods DP, Amasino RM (2017) Winter memory throughout the plant kingdom: different paths to flowering. Plant Physiol 173:27–35
Chen S, Songkumarn P, Liu J, Wang GL (2009) A versatile zero background T-vector system for gene cloning and functional genomics. Plant Physiol 150:1111–1121
Chen T, Cui P, Chen H, Ali S, Zhang S, Xiong L (2013) A KH-domain RNA-binding protein interacts with FIERY2/CTD phosphatase-like 1 and splicing factors and is important for pre-mRNA splicing in Arabidopsis. PLoS Genet 9(10):e1003875
Cheng Y, Kato N, Wang W, Li J, Chen X (2003) Two RNA binding proteins, HEN4 and HUA1, act in the processing of AGAMOUS pre-mRNA in Arabidopsis thaliana. Dev Cell 4:53–66
Clough SJ, Bent AF (1998) Floral dip: A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J 16:735–743
Csorba T, Questa JI, Sun Q, Dean C (2014) Antisense COOLAIR mediates the coordinated switching of chromatin states at FLC during vernalization. Proc Natl Acad Sci U S A 111:16160–16165
Dai G, Yin J, Li K, Chen D, Liu Z, Bi F, Rong C, Yao N (2020) The Arabidopsis AtGCD3 protein is a glucosylceramidase that preferentially hydrolyzes long-acyl-chain glucosylceramides. J Biol Chem 295(3):717–728
Dong C, Hu X, Tang W, Zheng X, Kim YS, Lee B, Zhu J (2006) A putative Arabidopsis nucleoporin, AtNUP160, is critical for RNA export and required for plant tolerance to cold stress. Mol Cell Biol 26(24):9533–9543
Fujiwara S, Oda A, Yoshida R, Niinuma K, Miyata K, Tomozoe Y, Tajima T, Nakagawa M, Hayashi K, Coupland G, Mizoguchi T (2008) Circadian clock proteins LHY and CCA1 regulate SVP protein accumulation to control flowering in Arabidopsis. Plant Cell 20:2960–2971
Gelvin SB (2017) Integration of Agrobacterium T-DNA into the plant genome. Annu Rev Genet 51:195–217
Grant JJ, Chini A, Basu D, Loake GJ (2003) Targeted activation tagging of the Arabidopsis NBS-LRR gene, ADR1, conveys resistance to virulent pathogens. Mol Plant Microbe Interact 16:669–680
Gu XF, Le C, Wang YZ, Li ZC, Jiang DH, Wang YQ, He YH (2013) Arabidopsis FLC clade members form flowering-repressor complexes coordinating responses to endogenous and environmental cues. Nat Commun 4:1947
Helliwell CA, Wood CC, Robertson M, James Peacock W, Dennis ES (2006) The Arabidopsis FLC protein interacts directly in vivo with SOC1 and FT chromatin and is part of a high-molecular-weight protein complex. Plant J 46:183–192
Hepworth SR (2002) Antagonistic regulation of flowering-time gene SOC1 by CONSTANS and FLC via separate promoter motifs. EMBO J 21:4327–4337
Johanson U, West J, Lister C, Michaels S, Amasino R, Dean C (2000) Molecular analysis of FRIGIDA, a major determinant of natural variation in Arabidopsis flowering time. Science 290:344–347
Jupe F, Rivkin AC, Michael TP, Zander M, Motley ST, Sandoval JP, Slotkin RK, Chen HM, Castanon R, Nery JR, Ecker JR (2019) The complex architecture and epigenomic impact of plant T-DNA insertions. PLoS Genet 15(1):e1007819
Kim DH, Sung S (2010) The plant homeo domain finger protein, VIN3-LIKE 2, is necessary for photoperiod-mediated epigenetic regulation of the floral repressor, MAF5. Proc Natl Acad Sci U S A 107:17029–17034
Kim DH, Sung S (2014) Genetic and epigenetic mechanisms underlying vernalization. Arabidopsis Book 12:e0171
Kosugi S, Hasebe M, Tomita M, Yanagawa H (2009) Systematic identification of yeast cell cycle-dependent nucleocytoplasmic shuttling proteins by prediction of composite motifs. Proc Natl Acad Sci U S A 106:10171–10176
Li H, Durbin R (2009) Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25(14):1754–1760
Lim MH, Kim J, Kim YS, Chung KS, Seo YH, Lee I, Hong CB, Kim HJ, Park CM (2004) A new Arabidopsis gene, FLK, encodes an RNA binding protein with K homology motifs and regulates flowering time via FLOWERINGLOCUS C. Plant Cell 16:731–740
Liu F, Quesada V, Crevill En P, Bäurle I, Swiezewski S, Dean C (2007) The arabidopsis RNA-binding protein FCA requires a lysinespecific demethylase 1 homolog to downregulate FLC. Mol Cell 28:398–407
Liu F, Marquardt S, Lister C, Swiezewski S, Dean C (2010) Targeted 3’ processing of antisense transcripts trggers arabidopsis FLC chromatin silencing. Science 327:94–97
Lorkovic ZJ (2009) Role of plant RNA-binding proteins in development, stress response and genome organization. Trends Plant Sci 14:229–236
Lorkovic ZJ, Barta A (2002) Genome analysis: RNA recognition motif (RPM) and K homology (KH) domain RNA-binding proteins from the flowering plant Arabidopsis thaliana. Nucleic Acids Res 30:623–635
Marquardt S, Raitskin O, Wu Z, Liu F, Sun Q, Dean C (2014) Functional consequences of splicing of the antisense transcript COOLAIR on FLC transcription. Mol Cell 54:156–165
Michaels S, Amasino R (1999) FLOWERING LOCUS C encodes a novel MADS domain protein that acts as a repressor of flowering. Plant Cell 11:949–956
Mockler TC, Yu X, Shalitin D, Parikh D, Michael TP, Liou J, Huang J, Smith Z, Alonso JM, Ecker JR, Chory J, Lin C (2004) Regulation of flowering time in Arabidopsis by K homology domain proteins. Proc Natl Acad Sci U S A 101:12759–12764
Ortuno-Miquel S, Rodriguez-Cazorla E, Zavala-Gonzalez EA, Martinez-Laborda A, Vera A (2019) Arabidopsis HUA ENHANCER 4 delays flowering by upregulating the MADS-box repressor genes FLC and MAF4. Sci Rep 9:1478
Parry G, Ward S, Cernac A, Dharmasiri S, Estelle M (2006) The Arabidopsis SUPPRESSOR OF AUXIN RESISTANCE proteins are nucleoporins with an important role in hormone signaling and development. Plant Cell 18:1590–1603
Ratcliffe OJ, Kumimoto RW, Wong BJ, Riechmann JL (2003) Analysis of the Arabidopsis MADS AFFECTING FLOWERING gene family: MAF2 prevents vernalization by short periods of cold. Plant Cell 15:1159–1169
Ripoll JJ, Rodríguez-Cazorla E, González-Reig S, Andújar A, Alonso-Cantabrana H, Perez-Amador MA, Carbonell J, Martínez-Laborda A, Vera A (2009) Antagonistic interactions between Arabidopsis K-homology domain genes uncover PEPPER as a positive regulator of the central floral repressor FLOWERING LOCUS C. Dev Biol 333:251–262
Rodriguez-Cazorla E, Ripoll JJ, Andujar A, Bailey LJ, Martinez-Laborda A, Yanofsky MF, Vera A (2015) K-homology nuclear ribonucleoproteins regulate floral organ identity and determinacy in Arabidopsis. PLoS Genet 11:2
Schmid M, Davison TS, Henz SR, Pape UJ, Demar M, Vingron M, Scholkopf B, Weigel D, Lohmann JU (2005) A gene expression map of Arabidopsis thaliana development. Nat Gen 37:501–506
Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D (2006) Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell 18:1121–1133
Scortecci KC, Michaels SD, Amasino RM (2001) Identification of a MADS-box gene, FLOWERING LOCUS M, that represses flowering. Plant J 26:229–236
Sheldon CC, Burn JE, Perez PP, Metzger J, Edwards JA, Peacock WJ, Dennis ES (1999) The FLF MADS box gene: a repressor of flowering in Arabidopsis regulated by vernalization and methylation. Plant Cell 11:445–458
Swiezewski S, Liu F, Magusin A, Dean C (2009) Cold-induced silencing by long antisense transcripts of an Arabidopsis Polycomb target. Nature 462:799–802
Thatcher LF, Kamphuis LG, Hane JK, Oñate-Sánchez L, Singh KB (2015) The Arabidopsis KH-domain RNA-binding protein ESR1 functions in components of jasmonate signalling, unlinking growth restraint and resistance to stress. PLoS ONE 10(5):e0126978
Thorvaldsdottir H, Robinson J, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14(2):178–192
Wang ZW, Wu Z, Raitskin Q, Sun Q, Dean C (2014) Antisense-mediated FLC transcriptional repression requires the P-TEFb transcription elongation factor. Proc Natl Acad Sci U S A 111:7468–7473
Werner JD, Borevitz JQ, Warthmann N, Trainer GT, Ecker JR, Chory J, Weigel D (2005) Quantitative trait locus mapping and DNA array hybridization identify an FLM deletion as a cause for natural flowering-time variation. Proc Natl Acad Sci U S A 102:2460–2465
Wu JX, Li J, Liu Z, Yin J, Chang ZY, Rong C, Wu JL, Bi FC, Yao N (2015) The Arabidopsis ceramidase AtACER functions in disease resistance and salt tolerance. Plant J 81:767–780
Xiao R, Chen J, Liang Z, Luo D, Chen G, Lu ZJ, Chen Y, Zhou B, Li H, Du X, Yang Y, San M, Wei X, Liu W, Lecuyer E, Graveley BR, Yeo GW, Burge CB, Zhang MQ, Zhou Y, Fu X (2019) Pervasive chromatin-RNA binding protein interactions enable RNA-based regulation of transcription. Cell 178:107–121.e18
Xing D, Zhao H, Xu R, Li QQ (2008) Arabidopsis PCFS4, a homolog of yeast polyadenylation factor Pcf11p, regulates FCA alternative processing and promotes flowering time. Plant J 54:899–910
Acknowledgements
We would like to thank Dr. Tao Chen (Sun Yat-sen University) for discussions about the manuscript, Dr. Detlef Weigel for the kind gift of the pRS300 vector, and the Arabidopsis Biological Resource Center for providing the Arabidopsis T-DNA insertion lines. This work was supported by the National Natural Science Foundation of China (31771357), the Natural Science Foundation of Guangdong Province (2017A030311005), and the Open Funding of Guangdong Provincial Key Laboratory of Plant Resource (PlantKF06).
Author information
Authors and Affiliations
Contributions
GYD and NY conceived and designed research. GYD, DKC, YPS, WYL, LQH, YKL and JFH conducted experiments. YL performed the initial NGS data analysis. GYD and NY wrote the manuscript. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by Neal Stewart.
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Dai, GY., Chen, DK., Sun, YP. et al. The Arabidopsis KH-domain protein FLOWERING LOCUS Y delays flowering by upregulating FLOWERING LOCUS C family members. Plant Cell Rep 39, 1705–1717 (2020). https://doi.org/10.1007/s00299-020-02598-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-020-02598-w