Abstract
Key message
Auxin and two phytochrome-interacting factors, PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and PIF5, play crucial roles in the enhancement of hypocotyl elongation in transgenic Arabidopsis thaliana plants that overproduce LOV KELCH PROTEIN2 (LKP2).
Abstract
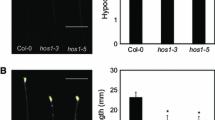
LOV KELCH PROTEIN2 (LKP2) is a positive regulator of hypocotyl elongation under white light in Arabidopsis thaliana. In this study, using microarray analysis, we compared the gene expression profiles of hypocotyls of wild-type Arabidopsis (Columbia accession), a transgenic line that produces green fluorescent protein (GFP), and two lines that produce GFP-tagged LKP2 (GFP-LKP2). We found that, in GFP-LKP2 hypocotyls, 775 genes were up-regulated, including 36 auxin-responsive genes, such as 27 SMALL AUXIN UP RNA (SAUR) and 6 AUXIN/INDOLE-3-ACETIC ACID (AUX/IAA) genes, and 21 genes involved in responses to red or far-red light, including PHYTOCHROME-INTERACTING FACTOR4 (PIF4) and PIF5; and 725 genes were down-regulated, including 15 flavonoid biosynthesis genes. Hypocotyls of GFP-LKP2 seedlings, but not cotyledons or roots, contained a higher level of indole-3-acetic acid (IAA) than those of control seedlings. Auxin inhibitors reduced the enhancement of hypocotyl elongation in GFP-LKP2 seedlings by inhibiting the increase in cortical cell number and elongation of the epidermal and cortical cells. The enhancement of hypocotyl elongation was completely suppressed in progeny of the crosses between GFP-LKP2 lines and dominant gain-of-function auxin-resistant mutants (axr2-1 and axr3-1) or loss-of-function mutants pif4, pif5, and pif4 pif5. Our results suggest that the enhancement of hypocotyl elongation in GFP-LKP2 seedlings is due to the elevated level of IAA and to the up-regulated expression of PIF4 and PIF5 in hypocotyls.






Similar content being viewed by others
References
Bae G, Choi G (2008) Decoding of light signals by plant phytochromes and their interacting proteins. Annu Rev Plant Biol 59:281–311
Baudry A, Ito S, Song YH, Strait AA, Kiba T, Lu S, Henriques R, Pruneda-Paz JL, Chua N-H, Tobin EM, Kay SA, Imaizumi T (2010) F-box proteins FKF1 and LKP2 act in concert with ZEITLUPE to control Arabidopsis clock progression. Plant Cell 22:606–622
Bauer D, Viczián A, Kircher S, Nobis T, Nitschke R, Kunkel T, Panigrahi KC, Adám E, Fejes E, Schäfer E, Nagy F (2004) Constitutive photomorphogenesis 1 and multiple photoreceptors control degradation of phytochrome interacting factor 3, a transcription factor required for light signaling in Arabidopsis. Plant Cell 16:1433–1445
Bernasconi P, Patel BC, Reagan JD, Subramanian MV (1996) The N-1-naphthylphthalamic acid-binding protein is an integral membrane protein. Plant Physiol 111:427–432
Boron AK, Vissenberg K (2014) The Arabidopsis thaliana hypocotyl, a model to identify and study control mechanisms of cellular expansion. Plant Cell Rep 33:697–706
Castillon A, Shen H, Huq E (2007) Phytochrome interacting factors: central players in phytochrome-mediated light signaling networks. Trends Plant Sci 12:514–521
Chae K, Isaacs CG, Reeves PH, Maloney GS, Muday GK, Nagpal P, Reed JW (2012) Arabidopsis SMALL AUXIN UP RNA63 promotes hypocotyl and stamen filament elongation. Plant J 71:684–697
Chapman EJ, Greenham K, Castillejo C, Sartor R, Bialy A, Sun TP, Estelle M (2012) Hypocotyl transcriptome reveals auxin regulation of growth-promoting genes through GA-dependent and -independent pathways. PLoS One 7:e36210
Cowling RJ, Harberd NP (2007) Gibberellins control Arabidopsis hypocotyl growth via regulation of cellular elongation. J Exp Bot 58:4269–4281
de Lucas M, Prat S (2014) PIFs get BRright: PHYTOCHROME INTERACTING FACTORs as integrators of light and hormonal signals. New Phytol 202:1126–1141
Demarsy E, Fankhauser C (2009) Higher plants use LOV to perceive blue light. Curr Opin Plant Biol 12:69–74
Dowson-Day MJ, Millar AJ (1999) Circadian dysfunction causes aberrant hypocotyl elongation patterns in Arabidopsis. Plant J 17:63–71
Duek PD, Fankhauser C (2005) bHLH class transcription factors take centre stage in phytochrome signalling. Trends Plant Sci 10:51–54
Fornara F, de Montaigu A, Sánchez-Villarreal A, Takahashi Y, Loren Ver, van Themaat E, Huettel B, Davis SJ, Coupland G (2015) The GI-CDF module of Arabidopsis affects freezing tolerance and growth as well as flowering. Plant J 81:695–706
Franklin KA, Lee SH, Patel D, Kumar SV, Spartz AK, Gu C, Ye S, Yu P, Breen G, Cohen JD, Wigge PA, Gray WM (2011) Phytochrome-interacting factor 4 (PIF4) regulates auxin biosynthesis at high temperature. Proc Natl Acad Sci USA 108:20231–20235
Hagen G, Guilfoyle T (2002) Auxin-responsive gene expression: genes, promoters and regulatory factors. Plant Mol Biol 49:373–385
Hayashi K, Neve J, Hirose M, Kuboki A, Shimada Y, Kepinski S, Nozaki H (2012) Rational design of an auxin antagonist of the SCF(TIR1) auxin receptor complex. ACS Chem Biol 7:590–598
Hornitschek P, Kohnen MV, Lorrain S, Rougemont J, Ljung K, López-Vidriero I, Franco-Zorrilla JM, Solano R, Trevisan M, Pradervand S, Xenarios I, Fankhauser C (2012) Phytochrome interacting factors 4 and 5 control seedling growth in changing light conditions by directly controlling auxin signaling. Plant J 71:699–711
Huang DW, Sherman BT, Lempicki RA (2009a) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nature Protoc 4:44–57
Huang DW, Sherman BT, Lempicki RA (2009b) Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res 37:1–13
Huang W, Pérez-García P, Pokhilko A, Millar AJ, Antoshechkin I, Riechmann JL, Más P (2012) Mapping the core of the Arabidopsis circadian clock defines the network structure of the oscillator. Science 336:75–79
Jarillo JA, Capel J, Tang RH, Yang HQ, Alonso JM, Ecker JR, Cashmore AR (2001) An Arabidopsis circadian clock component interacts with both CRY1 and phyB. Nature 410:487–490
Jiao Y, Lau OS, Deng XW (2007) Light-regulated transcriptional networks in higher plants. Nat Rev Genet 8:217–230
Kendrick RE, Kronenberg GHM (1994) Photomorphogenesis in plants. Kluwer Academic Publishers, Dordrecht, Boston, London
Kiba T, Henriques R, Sakakibara H, Chua N-H (2007) Targeted degradation of PSEUDO-RESPONSE REGULATOR5 by SCFZTL complex regulates clock function and photomorphogenesis in Arabidopsis thaliana. Plant Cell 19:2516–2530
Kiyosue T, Wada M (2000) LKP1 (LOV Kelch protein1): a factor involved in the regulation of flowering time in Arabidopsis. Plant J 23:807–815
Koini MA, Alvey L, Allen T, Tilley CA, Harberd NP, Whitelam GC, Franklin KA (2009) High temperature-mediated adaptations in plant architecture require the bHLH transcription factor PIF4. Curr Biol 19:408–413
Lau OS, Deng XW (2010) Plant hormone signaling lightens up: integrators of light and hormones. Curr Opin Plant Biol 13:571–577
Lau OS, Deng XW (2012) The photomorphogenic repressors COP1 and DET1: 20 years later. Trends Plant Sci 17:584–593
Lorrain S, Allen T, Duek PD, Whitelam GC, Fankhauser C (2008) Phytochrome-mediated inhibition of shade avoidance involves degradation of growth-promoting bHLH transcription factors. Plant J 53:312–323
Mancinelli AL (1990) Interaction between light quality and light quantity in the photoregulation of anthocyanin production. Plant Physiol 92:1191–1195
Más P, Kim WY, Somers DE, Kay SA (2003) Targeted degradation of TOC1 by ZTL modulates circadian function in Arabidopsis thaliana. Nature 426:567–570
Michael TP, Breton G, Hazen SP, Priest H, Mockler TC, Kay SA, Chory J (2008) A morning-specific phytohormone gene expression program underlying rhythmic plant growth. PLoS Biol 6:e225
Millar AJ (2004) Input signals to the plant circadian clock. J Exp Bot 55:277–283
Miyazaki Y, Yoshizumi T, Takase T, Matsui M, Kiyosue T (2011) Overexpression of LOV KELCH PROTEIN2 enhances cell elongation and increases cell number and ploidy in the hypocotyl of Arabidopsis thaliana. Plant Biotechnol 28:267–272
Mockler TC, Michael TP, Priest HD, Shen R, Sullivan CM, Givan SA, McEntee C, Kay SA, Chory J (2007) The DIURNAL project: DIURNAL and circadian expression profiling, model-based pattern matching, and promoter analysis. Cold Spring Harb Symp Quant Biol 72:353–363
Nagpal P, Walker LM, Young JC, Sonawala A, Timpte C, Estelle M, Reed JW (2000) AXR2 encodes a member of the Aux/IAA protein family. Plant Physiol 123:563–574
Nakamichi N, Kiba T, Henriques R, Mizuno T, Chua N-H, Sakakibara H (2010) PSEUDO-RESPONSE REGULATORS 9, 7, and 5 are transcriptional repressors in the Arabidopsis circadian clock. Plant Cell 22:594–605
Nelson DC, Lasswell J, Rogg LE, Cohen MA, Bartel B (2000) FKF1, a clock-controlled gene that regulates the transition to flowering in Arabidopsis. Cell 101:331–340
Nozue K, Maloof JN (2006) Diurnal regulation of plant growth. Plant Cell Environ 29:396–408
Nozue K, Covington MF, Duek PD, Lorrain S, Fankhauser C, Harmer SL, Maloof JN (2007) Rhythmic growth explained by coincidence between internal and external cues. Nature 448:358–361
Nozue K, Harmer SL, Maloof JN (2011) Genomic analysis of circadian clock-, light-, and growth-correlated genes reveals PHYTOCHROME-INTERACTING FACTOR5 as a modulator of auxin signaling in Arabidopsis. Plant Physiol 156:357–372
Nusinow DA, Helfer A, Hamilton EE, King JJ, Imaizumi T, Schultz TF, Farré EM, Kay SA (2011) The ELF4-ELF3-LUX complex links the circadian clock to diurnal control of hypocotyl growth. Nature 475:398–402
Oono Y, Ooura C, Rahman A, Aspuria ET, Hayashi K, Tanaka A, Uchimiya H (2003) p-Chlorophenoxyisobutyric acid impairs auxin response in Arabidopsis root. Plant Physiol 133:1135–1147
Roig-Villanova I, Bou-Torrent J, Galstyan A, Carretero-Paulet L, Portolés S, Rodríguez-Concepción M, Martínez-García JF (2007) Interaction of shade avoidance and auxin responses: a role for two novel atypical bHLH proteins. EMBO J 26:4756–4767
Saitoh A, Takase T, Kitaki H, Kiyosue T (2015a) Gene expression profile of Arabidopsis plants that overexpress ZEITLUPE/LOV KELCH PROTEIN1: up-regulation of auxin-inducible genes in hypocotyls. Plant Biotechnol. doi:10.5511/plantbiotechnology.15.0615b
Saitoh A, Takase T, Kitaki H, Miyazaki Y, Kiyosue T (2015b) Gene expression profile of zeitlupe/lov kelch protein1 T-DNA insertion mutants in Arabidopsis thaliana: downregulation of auxin-inducible genes in hypocotyls. Plant Signal Behav 10:e1071752
Schultz TF, Kiyosue T, Yanovsky M, Wada M, Kay SA (2001) A role for LKP2 in the circadian clock of Arabidopsis. Plant Cell 13:2659–2670
Shikata H, Hanada K, Ushijima T, Nakashima M, Suzuki Y, Matsushita T (2014) Phytochrome controls alternative splicing to mediate light responses in Arabidopsis. Proc Natl Acad Sci USA 111:18781–18786
Somers DE, Schultz TF, Milnamow M, Kay SA (2000) ZEITLUPE encodes a novel clock-associated PAS protein from Arabidopsis. Cell 101:319–329
Somers DE, Kim WY, Geng R (2004) The F-box protein ZEITLUPE confers dosage-dependent control on the circadian clock, photomorphogenesis, and flowering time. Plant Cell 16:769–782
Spartz AK, Lee SH, Wenger JP, Gonzalez N, Itoh H, Inzé D, Peer WA, Murphy AS, Overvoorde PJ, Gray WM (2012) The SAUR19 subfamily of SMALL AUXIN UP RNA genes promote cell expansion. Plant J 70:978–990
Spartz AK, Ren H, Park MY, Grandt KN, Lee SH, Murphy AS, Sussman MR, Overvoorde PJ, Gray WM (2014) SAUR inhibition of PP2C-D phosphatases activates plasma membrane H+-ATPases to promote cell expansion in Arabidopsis. Plant Cell 26:2129–2142
Staswick PE, Serban B, Rowe M, Tiryaki I, Maldonado MT, Maldonado MC, Suza W (2005) Characterization of an Arabidopsis enzyme family that conjugates amino acids to indole-3-acetic acid. Plant Cell 17:616–627
Takase T, Nishiyama Y, Tanihibashi H, Ogura Y, Miyazaki Y, Yamada Y, Kiyosue T (2011) LOV KELCH PROTEIN2 and ZEITLUPE repress Arabidopsis photoperiodic flowering under non-inductive conditions, dependent on FLAVIN-BINDING KELCH REPEAT F-BOX1. Plant J 67:608–621
Takase T, Miyazaki Y, Yasuhara M, Mitsui S, Kiyosue T (2015) Pleiotropic phenotype of transgenic Arabidopsis plants that produce the LOV domain of LOV KELCH PROTEIN2 (LKP2). Plant Biotechnol doi:10.5511/plantbiotechnology.15.0808b
Tanaka K, Nakamura Y, Asami T, Yoshida S, Matsuo T, Okamoto S (2003) Physiological roles of brassinosteroids in early growth of Arabidopsis: brassinosteroids have a synergistic relationship with gibberellin as well as auxin in light-grown hypocotyl elongation. J Plant Growth Regul 22:259–271
Tiwari SB, Wang XJ, Hagen G, Guilfoyle TJ (2001) AUX/IAA proteins are active repressors, and their stability and activity are modulated by auxin. Plant Cell 13:2809–2822
Valvekens D, Van Montagu M, Van Lijsebettens M (1988) Agrobacterium tumefaciens-mediated transformation of Arabidopsis thaliana root explants by using kanamycin selection. Proc Natl Acad Sci USA 85:5536–5540
Vandenbussche F, Verbelen JP, van der Straeten D (2005) Of light and length: regulation of hypocotyl growth in Arabidopsis. BioEssays 27:275–284
Woodward AW, Bartel B (2005) Auxin: regulation, action, and interaction. Ann Bot 95:707–735
Yasuhara M, Mitsui S, Hirano H, Takanabe R, Tokioka Y, Ihara N, Komatsu A, Seki M, Shinozaki K, Kiyosue T (2004) Identification of ASK and clock-associated proteins as molecular partners of LKP2 (LOV kelch protein 2) in Arabidopsis. J Exp Bot 55:2015–2027
Yoshimoto K, Jikumaru Y, Kamiya Y, Kusano M, Consonni C, Panstruga R, Ohsumi Y, Shirasu K (2009) Autophagy negatively regulates cell death by controlling NPR1-dependent salicylic acid signaling during senescence and the innate immune response in Arabidopsis. Plant Cell 21:2914–2927
Zoltowski BD, Imaizumi T (2014) Structure and function of the ZTL/FKF1/LKP2 group proteins in Arabidopsis. Enzymes 35:213–239
Acknowledgments
This research was partially supported by a grant from the Ministry of Education, Culture, Sports, Science and Technology of Japan. We thank Dr. K. Hayashi (Okayama University of Science, Okayama, Japan) for kindly providing PEO-IAA, Dr. J. N. Maloof (University of California, Davis, CA, USA) for kindly providing pif4 and pif5 seeds, and ABRC for providing axr2-1 and axr3-1 seeds.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no conflict of interest.
Additional information
Communicated by P. P. Kumar.
Electronic supplementary material
Below is the link to the electronic supplementary material.
299_2015_1896_MOESM4_ESM.xlsx
Table S4. Pathway analysis of genes up-regulated in GFP-LKP2 hypocotyls performed with the Gene Functional Classification Tool in DAVID Bioinformatics Resources 6.7 (XLSX 20 kb)
299_2015_1896_MOESM5_ESM.xlsx
Table S5. Pathway analysis of genes down-regulated in GFP-LKP2 hypocotyls performed with the Gene Functional Classification Tool in DAVID Bioinformatics Resources 6.7 (XLSX 22 kb)
299_2015_1896_MOESM6_ESM.tif
Supplemental Fig. S1. Embryos of control (Col and GFP) and GFP-LKP2 (lines GFP-LKP2-1 and GFP-LKP2-2). (A) Embryos were surgically excised from 6-h water-imbibed seeds and stained with 0.1 % (w/v) Toluidine blue. Bars = 250 μm (B) Epidermal cell number on a 250-μm line along the long axis of the hypocotyl at its thickest part were counted. Error bars represent standard error of the mean (n = 5–10); the same letters indicate no statistically significant differences (P ≥ 0.05, Tukey’s test) (TIFF 20705 kb)
299_2015_1896_MOESM7_ESM.tif
Supplemental Fig. S2. Seedling growth. (A) Control (Col and GFP) and GFP-LKP2 (lines GFP-LKP2-1 and GFP-LKP2-2) seeds and seedlings grown on the germination medium for 0 to 7 days under continuous white light (80 μmol·m−2·s−1) at 22 °C. (B) Time course of hypocotyl elongation (TIFF 11131 kb)
299_2015_1896_MOESM8_ESM.jpg
Supplemental Fig. S3. Five-day-old control (Col and GFP) and GFP-LKP2 seedlings (lines GFP-LKP2-1 and GFP-LKP2-2) in the absence or presence of indicated concentrations of N-1-naphthylphthalamic acid (NPA), p-chlorophenoxyisobutyric acid (PCIB), or α-(phenylethyl-2-one)-indole-3-acetic acid (PEO-IAA). Bar = 5 mm (JPEG 908 kb)
299_2015_1896_MOESM9_ESM.tif
Supplemental Fig. S4. Inhibition of hypocotyl elongation by auxin inhibitors. Hypocotyl length of 5-day-old control (Col and GFP) and GFP-LKP2 seedlings (lines GFP-LKP2-1 and GFP-LKP2-2) in the absence or presence of the indicated concentrations of N-1-naphthylphthalamic acid (NPA), p-chlorophenoxyisobutyric acid (PCIB), or α-(phenylethyl-2-one)-indole-3-acetic acid (PEO-IAA). Error bars represent standard error of the mean (n = 7–15); different letters indicate statistically significant differences (P < 0.05, Tukey’s test) (TIFF 5375 kb)
Rights and permissions
About this article
Cite this article
Miyazaki, Y., Jikumaru, Y., Takase, T. et al. Enhancement of hypocotyl elongation by LOV KELCH PROTEIN2 production is mediated by auxin and phytochrome-interacting factors in Arabidopsis thaliana . Plant Cell Rep 35, 455–467 (2016). https://doi.org/10.1007/s00299-015-1896-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1896-4