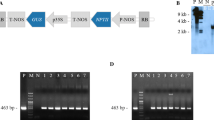
Abstract
Key message
We found 35S promoter sequence-specific DNA methylation in lettuce. Additionally, transgenic lettuce plants having a modified 35S promoter lost methylation, suggesting the modified sequence is subjected to the methylation machinery.
Abstract
We previously reported that cauliflower mosaic virus 35S promoter-specific DNA methylation in transgenic gentian (Gentiana triflora × G. scabra) plants occurs irrespective of the copy number and the genomic location of T-DNA, and causes strong gene silencing. To confirm whether 35S-specific methylation can occur in other plant species, transgenic lettuce (Lactuca sativa L.) plants with a single copy of the 35S promoter-driven sGFP gene were produced and analyzed. Among 10 lines of transgenic plants, 3, 4, and 3 lines showed strong, weak, and no expression of sGFP mRNA, respectively. Bisulfite genomic sequencing of the 35S promoter region showed hypermethylation at CpG and CpWpG (where W is A or T) sites in 9 of 10 lines. Gentian-type de novo methylation pattern, consisting of methylated cytosines at CpHpH (where H is A, C, or T) sites, was also observed in the transgenic lettuce lines, suggesting that lettuce and gentian share similar methylation machinery. Four of five transgenic lettuce lines having a single copy of a modified 35S promoter, which was modified in the proposed core target of de novo methylation in gentian, exhibited 35S hypomethylation, indicating that the modified sequence may be the target of the 35S-specific methylation machinery.





Similar content being viewed by others
Abbreviations
- CaMV:
-
Cauliflower mosaic virus
- sGFP:
-
Synthetic green fluorescent protein
- T-DNA:
-
Transfer DNA
References
Balàzs E, Lebeurier G (1981) Arabidopsis is a host of cauliflower mosaic virus. Arabidopsis Newslett 18:130–134
Benfey PN, Chua NH (1990) The cauliflower mosaic virus 35S promoter: combinatorial regulation of transcription in plants. Science 250:959–966
Butaye KMJ, Cammue BPA, Delauré SL, De Bolle MFC (2005) Approaches to minimize variation of transgene expression in plants. Mol Breed 16:79–91
Dan S, Qiang H, Zhaonan D, Zhengquan H (2014) Genetic transformation of lettuce (Lactuca sativa): a review. Afr J Biotechnol 13:1686–1693
Depicker A, Sanders M, Meyer P (2005) Transgene silencing. In: Meyer P (ed) Plant epigenetics, annual plant reviews, vol 19. Blackwell, Oxford, pp 1–32
Dong H, Zhao Y, Wang Y, Li H (2014) Recombinant proteins expressed in lettuce. Indian J Biotechnol 13:427–436
Finn TE, Wang L, Smolilo D, Smith NA, White R, Chaudhury A, Dennis ES, Wang MB (2011) Transgene expression and transgene-induced silencing in diploid and autotetraploid Arabidopsis. Genetics 187:409–423
Frommer M, McDonald LE, Millar DS, Collis CM, Watt F, Grigg GW, Molloy PL, Paul CL (1992) A genomic sequencing protocol that yields a positive display of 5-methylcytosine residues in individual DNA strands. Proc Natl Acad Sci USA 89:1827–1831
Gelvin SB (2000) Agrobacterium and plant genes involved in T-DNA transfer and integration. Annu Rev Plant Physiol Plant Mol Biol 51:223–256
Gelvin SB (2003) Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiol Mol Biol Rev 67:16–37
Hirai T, Shohael AM, Kim Y-W, Yano M, Ezura H (2011) Ubiquitin promoter–terminator cassette promotes genetically stable expression of the taste-modifying protein miraculin in transgenic lettuce. Plant Cell Rep 30:2255–2265
Höfgen R, Willmitzer L (1988) Storage of competent cells for Agrobacterium transformation. Nucleic Acids Res 16:9877
Hood EE, Helmer GL, Fraley RT, Chilton MD (1986) The hypervirulence of Agrobacterium tumefaciens A281 is encoded in a region of pTiBo542 outside of T-DNA. J Bacteriol 168:1291–1301
Horsch RB, Fry JE, Hoffmann NL, Eichholtz D, Rogers SG, Fraley RT (1985) A simple and general method for transferring genes into plants. Science 227:1229–1231
Imamura T, Nakatsuka T, Higuchi A, Nishihara M, Takahashi H (2011) The gentian orthologues of FT/TFL1 gene family control floral initiation in Gentiana. Plant Cell Physiol 52:1031–1041
Kuroda T, Urushibara S, Takeda I, Nakatani F, Suzuki K (2002) Multiplex reverse transcription polymerase chain reaction for simultaneous detection of viruses in gentian. J Gen Plant Pathol 68:169–172
Lam E, Benfey PN, Gilmartin PM, Fang R-X, Chua N-H (1989) Site-specific mutations alter in vitro factor binding and change promoter expression pattern in transgenic plants. Proc Natl Acad Sci USA 86:7890–7894
Matzke MA, Mette MF, Matzke AJM (2000) Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol 43:401–415
McCabe MS, Schepers F, van der Arend A, Mohapatra U, de Laat AMM, Power JB, Davey MR (1999) Increased stable inheritance of herbicide resistance in transgenic lettuce carrying a petE promoter-bar gene compared with a CaMV 35S-bar gene. Theor Appl Genet 99:587–592
Melcher U, Brannan CM, Gardner CO Jr, Essenberg RC (1992) Diverse mechanisms of plant resistance to cauliflower mosaic virus revealed by leaf skeleton hybridization. Arch Virol 123:379–387
Mishiba K-I, Nishihara M, Nakatsuka T, Abe Y, Hirano H, Yokoi T, Kikuchi A, Yamamura S (2005) Consistent transcriptional silencing of 35S-driven transgenes in gentian. Plant J 44:541–556
Mishiba K-I, Nishihara M, Abe Y, Nakatsuka T, Kawamura H, Kodama K, Takesawa T, Abe J, Yamamura S (2006) Production of dwarf potted gentian using wild-type Agrobacterium rhizogenes. Plant Biotechnol 23:33–38
Mishiba K-I, Yamasaki S, Nakatsuka T, Abe Y, Daimon H, Oda M, Nishihara M (2010) Strict de novo methylation of the 35S enhancer sequence in gentian. PLoS One 5:e9670
Murashige T, Skoog F (1962) A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol Plant 15:473–497
Nagaya S, Kato K, Ninomiya Y, Horie R, Sekine M, Yoshida K, Shinmyo A (2005) Expression of randomly integrated single copy transgenes does not vary in Arabidopsis thaliana. Plant Cell Physiol 46:438–444
Nakatsuka T, Mishiba K-I, Abe Y, Kubota A, Kakizaki Y, Yamamura S, Nishihara M (2008) Flower color modification of gentian plants by RNAi-mediated gene silencing. Plant Biotechnol 25:61–68
Nakatsuka T, Abe Y, Kakizaki Y, Kubota A, Shimada N, Nishihara M (2009) Over-expression of Arabidopsis FT gene reduces juvenile phase and induces early flowering in ornamental gentian plants. Euphytica 168:113–119
Nakatsuka T, Mishiba K-I, Kubota A, Abe Y, Yamamura S, Nakamura N, Tanaka Y, Nishihara M (2010) Genetic engineering of novel flower colour by suppression of anthocyanin modification genes in gentian. J Plant Physiol 167:231–237
Niwa Y, Hirano T, Yoshimoto K, Shimizu M, Kobayashi H (1999) Non-invasive quantitative detection and applications of non-toxic, S65T-type green fluorescent protein in living plants. Plant J 18:455–463
Scheid OM, Jakovleva L, Afsar K, Maluszynska J, Paszkowski J (1996) A change of ploidy can modify epigenetic silencing. Proc Natl Acad Sci USA 93:7114–7119
Schoelz JE, Shepherd RJ (1988) Host range control of cauliflower mosaic virus. Virology 162:30–37
Schoelz JE, Shepherd RJ, Richins RD (1986) Properties of an unusual strain of cauliflower mosaic virus. Phytopathology 76:451–454
Schubert D, Lechtenberg B, Forsbach A, Gils M, Bahadur S, Schmidt R (2004) Silencing in Arabidopsis T-DNA transformants: the predominant role of a gene-specific RNA sensing mechanism versus position effects. Plant Cell 16:2561–2572
Sun H-J, Cui M-L, Ma B, Ezura H (2006) Functional expression of the taste-modifying protein, miraculin, in transgenic lettuce. FEBS Lett 580:620–626
Thompson CJ, Movva NR, Tizard R, Crameri R, Davies JE, Lauwereys M, Botterman J (1987) Characterization of the herbicide-resistance gene bar from Streptomyces hygroscopicus. EMBO J 6:2519–2523
Yamasaki S, Oda M, Koizumi N, Mitsukuri K, Johkan M, Nakatsuka T, Nishihara M, Mishiba K-I (2011a) De novo DNA methylation of the 35S enhancer revealed by high-resolution methylation analysis of an entire T-DNA segment in transgenic gentian. Plant Biotechnol 28:223–230
Yamasaki S, Oda M, Daimon H, Mitsukuri K, Johkan M, Nakatsuka T, Nishihara M, Mishiba K-I (2011b) Epigenetic modifications of the 35S promoter in cultured gentian cells. Plant Sci 180:612–619
Yano M, Hirai T, Kato K, Hiwasa-Tanase K, Fukuda N, Ezura H (2010) Tomato is a suitable material for producing recombinant miraculin protein in genetically stable manner. Plant Sci 178:469–473
Acknowledgments
We thank Ms. Ayumi Sakei (Osaka Prefecture University) for technical assistance. This work was supported by a Grant-in-Aid for Scientific Research (Grant Number, 23780027) from The Ministry of Education, Culture, Sports, Science and Technology (MEXT).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Communicated by K. K. Kamo.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Okumura, A., Shimada, A., Yamasaki, S. et al. CaMV-35S promoter sequence-specific DNA methylation in lettuce. Plant Cell Rep 35, 43–51 (2016). https://doi.org/10.1007/s00299-015-1865-y
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00299-015-1865-y